Legend
| %id | Code | Chains | Pic | Error? | Sym | Nsub | Sprot | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % seq. id. | PDB Code | query & target | Graph picture | Is the current PDB state judged to be an error? | Symmetry | No. subunits | SwissProt link | No. of amino acids (per chain) | Pubmed ID | Get Homologs | Edit Structure | View structure | Organism | Function |
Protein reference
| Code | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto? | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB | PiQSi | |||||||||||||||||
| 4xi1_3 |  | ? | NPS | 1 | ? | ? | - | - | - | Q5X159 | P | H | E | R | Legionella pneumophila (strain Paris) | LIGASE | ||
Homologs
| %id | Code | Chains | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 4xi1_1 | C & A |  | ? | NPS | 1 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila (strain Paris) | LIGASE | |||
| 100 | 4xi1_2 | C & B |  | ? | NPS | 1 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila (strain Paris) | LIGASE | |||
| 100 | 4xi1_4 | C & E |  | ? | D3 | 6 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila (strain Paris) | LIGASE | |||
| 99 | 4wz2_1 | C & A |  | ? | NPS | 1 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila | LIGASE | |||
| 99 | 4wz2_2 | C & B |  | ? | NPS | 1 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila | LIGASE | |||
| 99 | 4wz2_3 | C & C |  | ? | NPS | 1 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila | LIGASE | |||
| 35 | 2oxq_1 | C & C |  | ? | C2 | 4 | ? | ? | ? | Q6PC58 | P | H | E | R | Danio rerio | Ligase | |||
| 32 | 2f42_1 | C & A | 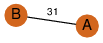 | ? | C2 | 2 | ? | ? | NO | C2 | 2 | Q7ZTZ6 | P | H | E | R | Danio rerio | CHAPERONE | |
| 30 | 2bay_1 | C & A | 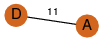 | ? | C2 | 2 | ? | ? | PROBNOT | C2 | 2 | P32523 | P | H | E | R | Saccharomyces cerevisiae | LIGASE | |
| 30 | 2bay_2 | C & C | 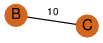 | ? | C2 | 2 | ? | ? | PROBNOT | C2 | 2 | P32523 | P | H | E | R | Saccharomyces cerevisiae | LIGASE | |
| 30 | 2bay_3 | C & F | 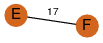 | ? | C2 | 2 | ? | ? | PROBNOT | C2 | 2 | P32523 | P | H | E | R | Saccharomyces cerevisiae | LIGASE | |
| 26 | 4wz0_1 | C & A |  | ? | NPS | 1 | ? | ? | ? | Q5X159 | P | H | E | R | Legionella pneumophila (strain Paris) | LIGASE | |||
| 25 | 3l1x_1 | C & A |  | ? | NPS | 1 | ? | ? | ? | O95155 | P | H | E | R | Homo sapiens | LIGASE | |||
| 25 | 3l1z_1 | C & B |  | ? | NPS | 2 | ? | ? | ? | P61077 | P | H | E | R | Homo sapiens | LIGASE | |||
| 25 | 3l1z_2 | C & B |  | ? | NPS | 2 | ? | ? | ? | P61077 | P | H | E | R | Homo sapiens | LIGASE | |||
| 0 | 2c2v_1 | C & T |  | ? | C2 | 6 | ? | ? | ? | 2C2V | P | H | E | R | MUS MUSCULUS | CHAPERONE | |||
| 0 | 2c2v_2 | C & V |  | ? | C2 | 6 | ? | ? | ? | 2C2V | P | H | E | R | MUS MUSCULUS | CHAPERONE |