Legend
| %id | Code | Chains | Pic | Error? | Sym | Nsub | Sprot | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % seq. id. | PDB Code | query & target | Graph picture | Is the current PDB state judged to be an error? | Symmetry | No. subunits | SwissProt link | No. of amino acids (per chain) | Pubmed ID | Get Homologs | Edit Structure | View structure | Organism | Function |
Protein reference
| Code | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto? | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB | PiQSi | |||||||||||||||||
| 4rxl_1 |  | ? | NPS | 1 | ? | ? | - | - | - | Q9KLL7 | P | H | E | R | Vibrio cholerae | TRANSPORT PROTEIN | ||
Homologs
| %id | Code | Chains | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 42 | 1amf_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | P37329 | P | H | E | R | 0 | BINDING PROTEIN | |||
| 42 | 1wod_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | P37329 | P | H | E | R | 0 | TRANSPORT | |||
| 42 | 3axf_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | P37329 | P | H | E | R | 0 | METAL BINDING PROTEIN | |||
| 42 | 3axf_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | P37329 | P | H | E | R | 0 | METAL BINDING PROTEIN | |||
| 42 | 3axf_3 | A & C |  | ? | NPS | 1 | ? | ? | ? | P37329 | P | H | E | R | 0 | METAL BINDING PROTEIN | |||
| 42 | 3r26_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | P37329 | P | H | E | R | Escherichia coli | PROTEIN BINDING | |||
| 41 | 2h5y_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL TRANSPORT | |||
| 41 | 2h5y_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL TRANSPORT | |||
| 41 | 2h5y_3 | A & C |  | ? | NPS | 1 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL TRANSPORT | |||
| 41 | 2h5y_4 | A & C |  | ? | C2 | 4 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL TRANSPORT | |||
| 41 | 2h5y_5 | A & A |  | ? | C2 | 2 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL TRANSPORT | |||
| 40 | 3gzg_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL BINDING PROTEIN | |||
| 40 | 3gzg_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL BINDING PROTEIN | |||
| 40 | 3gzg_3 | A & C |  | ? | NPS | 1 | ? | ? | ? | Q8PHA1 | P | H | E | R | Xanthomonas axonopodis pv. citri | METAL BINDING PROTEIN | |||
| 32 | 4kd5_1 | A & A |  | ? | C2 | 4 | ? | ? | ? | Q18A64 | P | H | E | R | Clostridium difficile | TRANSPORT PROTEIN | |||
| 32 | 4kd5_2 | A & C |  | ? | NPS | 1 | ? | ? | ? | Q18A64 | P | H | E | R | Clostridium difficile | TRANSPORT PROTEIN | |||
| 32 | 4kd5_3 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q18A64 | P | H | E | R | Clostridium difficile | TRANSPORT PROTEIN | |||
| 32 | 4kd5_4 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q18A64 | P | H | E | R | Clostridium difficile | TRANSPORT PROTEIN | |||
| 32 | 4kd5_5 | A & D |  | ? | NPS | 1 | ? | ? | ? | Q18A64 | P | H | E | R | Clostridium difficile | TRANSPORT PROTEIN | |||
| 24 | 1atg_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q7SIH2 | P | H | E | R | 0 | BINDING PROTEIN | |||
| 0 | 1sbp_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | P02906 | P | H | E | R | Salmonella typhimurium | BINDING PROTEIN | |||
| 0 | 2hxw_1 | A & B |  | ? | C2 | 2 | ? | ? | NO | C2 | 2 | Q9PIK7 | P | H | E | R | Campylobacter jejuni | periplasmic binding protein | |
| 0 | 2onk_1 | A & E |  | ? | NPS | 5 | ? | ? | ? | O30144 | P | H | E | R | Archaeoglobus fulgidus | MEMBRANE PROTEIN | |||
| 0 | 2onk_2 | A & J |  | ? | NPS | 5 | ? | ? | ? | O30144 | P | H | E | R | Archaeoglobus fulgidus | MEMBRANE PROTEIN | |||
| 0 | 2onr_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | O30142 | P | H | E | R | Archaeoglobus fulgidus DSM 4304 | LIGAND BINDING PROTEIN | |||
| 0 | 2ons_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | O30142 | P | H | E | R | Archaeoglobus fulgidus DSM 4304 | LIGAND BINDING PROTEIN | |||
| 0 | 3cfx_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 3cfx_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 3cfz_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q58586 | P | H | E | R | Methanocaldococcus jannaschii | TRANSPORT PROTEIN | |||
| 0 | 3cg1_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8U4K5 | P | H | E | R | Pyrococcus furiosus | TRANSPORT PROTEIN | |||
| 0 | 3cg1_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q8U4K5 | P | H | E | R | Pyrococcus furiosus | TRANSPORT PROTEIN | |||
| 0 | 3cg3_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | O57890 | P | H | E | R | Pyrococcus horikoshii | TRANSPORT PROTEIN | |||
| 0 | 3cij_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | O30142 | P | H | E | R | Archaeoglobus fulgidus | TRANSPORT PROTEIN | |||
| 0 | 3cij_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | O30142 | P | H | E | R | Archaeoglobus fulgidus | TRANSPORT PROTEIN | |||
| 0 | 3fir_1 | A & A |  | ? | C2 | 2 | ? | ? | NO | C2 | 2 | Q0PBL7 | P | H | E | R | Campylobacter jejuni | PROTEIN BINDING | |
| 0 | 3fj7_1 | A & B |  | ? | C2 | 2 | ? | ? | PROBNOT | C2 | 2 | Q0PBL7 | P | H | E | R | Campylobacter jejuni | PROTEIN BINDING | |
| 0 | 3fjg_1 | A & A |  | ? | C2 | 2 | ? | ? | NO | C2 | 2 | Q0PBL7 | P | H | E | R | Campylobacter jejuni | TRANSPORT PROTEIN | |
| 0 | 3fjg_2 | A & C |  | ? | C2 | 2 | ? | ? | NO | C2 | 2 | Q0PBL7 | P | H | E | R | Campylobacter jejuni | TRANSPORT PROTEIN | |
| 0 | 3fjm_1 | A & B | 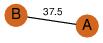 | ? | C2 | 2 | ? | ? | PROBNOT | C2 | 2 | Q0PBL7 | P | H | E | R | Campylobacter jejuni | TRANSPORT PROTEIN | |
| 0 | 3k6u_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 3k6v_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 3k6w_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 3k6x_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 3k6x_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q8TTZ5 | P | H | E | R | Methanosarcina acetivorans | TRANSPORT PROTEIN | |||
| 0 | 4jb7_1 | A & A | 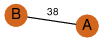 | ? | C2 | 2 | ? | ? | PROBNOT | C2 | 2 | Q9KTQ6 | P | H | E | R | Vibrio cholerae O1 | CELL INVASION |