Legend
| %id | Code | Chains | Pic | Error? | Sym | Nsub | Sprot | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % seq. id. | PDB Code | query & target | Graph picture | Is the current PDB state judged to be an error? | Symmetry | No. subunits | SwissProt link | No. of amino acids (per chain) | Pubmed ID | Get Homologs | Edit Structure | View structure | Organism | Function |
Protein reference
| Code | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto? | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB | PiQSi | |||||||||||||||||
| 3zix_5 |  | ? | NPS | 1 | ? | ? | - | - | - | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | ||
Homologs
| %id | Code | Chains | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 2quo_1 | E & A |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | Clostridium perfringens | Toxin | |||
| 100 | 3am2_1 | E & B |  | ? | C3 | 3 | ? | ? | ? | P01558 | P | H | E | R | Clostridium perfringens | TOXIN | |||
| 100 | 3zix_1 | E & A |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 100 | 3zix_2 | E & B |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 100 | 3zix_3 | E & C |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 100 | 3zix_4 | E & D |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 100 | 3zix_6 | E & F |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 100 | 4p5h_1 | E & K |  | ? | C3 | 3 | ? | ? | ? | O88552 | P | H | E | R | Clostridium perfringens | Toxin/CELL Adhesion | |||
| 100 | 4p5h_2 | E & N |  | ? | C3 | 3 | ? | ? | ? | O88552 | P | H | E | R | Clostridium perfringens | Toxin/CELL Adhesion | |||
| 100 | 4p5h_3 | E & A |  | ? | C3 | 3 | ? | ? | ? | O88552 | P | H | E | R | Clostridium perfringens | Toxin/CELL Adhesion | |||
| 100 | 4p5h_4 | E & F |  | ? | C3 | 3 | ? | ? | ? | O88552 | P | H | E | R | Clostridium perfringens | Toxin/CELL Adhesion | |||
| 100 | 4p5h_5 | E & G |  | ? | C3 | 3 | ? | ? | ? | O88552 | P | H | E | R | Clostridium perfringens | Toxin/CELL Adhesion | |||
| 99 | 2xh6_1 | E & A |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | 0 | TOXIN | |||
| 99 | 2xh6_2 | E & B |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | 0 | TOXIN | |||
| 99 | 2xh6_3 | E & C |  | ? | NPS | 1 | ? | ? | ? | P01558 | P | H | E | R | 0 | TOXIN | |||
| 99 | 2yhj_1 | E & A |  | ? | C3 | 3 | ? | ? | ? | P01558 | P | H | E | R | 0 | TOXIN | |||
| 99 | 2yhj_2 | E & A |  | ? | C3 | 3 | ? | ? | ? | P01558 | P | H | E | R | 0 | TOXIN | |||
| 99 | 3x29_1 | E & B |  | ? | Pb | 2 | ? | ? | ? | Q9ET38 | P | H | E | R | Mus musculus | CELL ADHESION | |||
| 99 | 3x29_2 | E & D |  | ? | Pb | 2 | ? | ? | ? | Q9ET38 | P | H | E | R | Mus musculus | CELL ADHESION | |||
| 99 | 3ziw_1 | E & A |  | ? | C3 | 3 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 99 | 3ziw_2 | E & E | 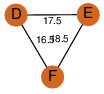 | ? | C3 | 3 | ? | ? | ? | P01558 | P | H | E | R | CLOSTRIDIUM PERFRINGENS | TOXIN | |||
| 26 | 4en6_1 | E & E |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | P46085 | P | H | E | R | Clostridium botulinum | SUGAR BINDING PROTEIN | |
| 26 | 4en7_1 | E & A |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | P46085 | P | H | E | R | Clostridium botulinum | SUGAR BINDING PROTEIN | |
| 26 | 4en8_1 | E & A |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | P46085 | P | H | E | R | Clostridium botulinum | SUGAR BINDING PROTEIN | |
| 26 | 4en9_1 | E & C |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | P46085 | P | H | E | R | Clostridium botulinum | SUGAR BINDING PROTEIN | |
| 26 | 2zoe_1 | E & E |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | P46085 | P | H | E | R | Clostridium botulinum | TOXIN | |
| 26 | 2zs6_1 | E & A |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | P46085 | P | H | E | R | Clostridium botulinum | TOXIN | |
| 23 | 3win_1 | E & L |  | ? | NPS | 15 | ? | ? | ? | Q33CP6 | P | H | E | R | Clostridium botulinum B | TOXIN | |||
| 23 | 4lo4_1 | E & A |  | ? | C6 | 6 | ? | ? | NO | C6 | 6 | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |
| 23 | 4lo5_1 | E & C |  | ? | C6 | 6 | ? | ? | PROBNOT | C6 | 6 | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |
| 23 | 4lo6_1 | E & C |  | ? | C6 | 6 | ? | ? | PROBNOT | C6 | 6 | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |
| 0 | 4lo7_1 | E & A |  | ? | NPS | 4 | ? | ? | ? | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |||
| 0 | 4lo7_2 | E & E |  | ? | NPS | 4 | ? | ? | ? | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |||
| 0 | 4lo7_3 | E & E |  | ? | NPS | 1 | ? | ? | PROBYES2 | C6 | 6 | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |
| 0 | 4lo7_5 | E & A |  | ? | NPS | 1 | ? | ? | PROBYES2 | C6 | 6 | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |
| 0 | 4lo8_1 | E & A | 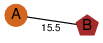 | ? | C2 | 2 | ? | ? | ? | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |||
| 0 | 4lo8_2 | E & C | 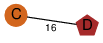 | ? | C2 | 2 | ? | ? | ? | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |||
| 0 | 4lo8_3 | E & E | 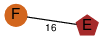 | ? | C2 | 2 | ? | ? | ? | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |||
| 0 | 4lo8_4 | E & G |  | ? | C2 | 2 | ? | ? | ? | Q8KHU9 | P | H | E | R | Clostridium botulinum | PROTEIN TRANSPORT | |||
| 0 | 4qd2_1 | E & A |  | ? | NPS | 5 | ? | ? | ? | A5HZZ4 | P | H | E | R | Clostridium botulinum | CELL ADHESION | |||
| 0 | 4qd2_2 | E & F |  | ? | NPS | 5 | ? | ? | ? | A5HZZ4 | P | H | E | R | Clostridium botulinum | CELL ADHESION |