to read PiQSi annotation, and roll over the PDB name to read its title.
| %id |
Code |
Chains |
Pic |
Error? |
Sym1 |
Nsub1 |
Sym2 |
Nsub2 |
Error_auto |
Sym_auto |
Nsub_auto |
SProt |
Weight |
Ref |
H |
E |
R |
Org |
Fun |
| 100 | 3m6n_1 | A & A | 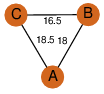 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q7CLS3 | | P | H | E | R | Xanthomonas campestris pv. campestris | LYASE |
| 36 | 1ab5_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 36 | 1ab5_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 36 | 1ab6_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 36 | 1ab6_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1a0o_1 | D & A |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1a0o_2 | D & C |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1a0o_3 | D & E |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1a0o_4 | D & G |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1bdj_1 | D & A |  | PROBYES
| NPS | 2 | NPS | 1 | ? |
| | P06143 | | P | H | E | R | Escherichia coli | COMPLEX (CHEMOTAXIS/TRANSFERASE) |
| 35 | 1chn_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNAL TRANSDUCTION PROTEIN |
| 35 | 1eay_1 | D & A |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | 0 | SIGNAL TRANSDUCTION COMPLEX |
| 35 | 1eay_2 | D & B |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | 0 | SIGNAL TRANSDUCTION COMPLEX |
| 35 | 1f4v_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1f4v_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1f4v_3 | D & C |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1ffg_1 | D & A |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | TRANSFERASE/SIGNALING PROTEIN |
| 35 | 1ffg_2 | D & C |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | TRANSFERASE/SIGNALING PROTEIN |
| 35 | 1ffs_1 | D & A |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | TRANSFERASE/SIGNALING PROTEIN |
| 35 | 1ffs_2 | D & C |  | ?
| NPS | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | TRANSFERASE/SIGNALING PROTEIN |
| 35 | 1ffw_1 | D & A |  | ?
| NPS | 2 | ? | ? | ? |
| | P07363 | | P | H | E | R | Escherichia coli | TRANSFERASE/SIGNALING PROTEIN |
| 35 | 1ffw_2 | D & C |  | ?
| NPS | 2 | ? | ? | ? |
| | P07363 | | P | H | E | R | Escherichia coli | TRANSFERASE/SIGNALING PROTEIN |
| 35 | 1fqw_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1fqw_2 | D & B |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1jbe_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1kmi_1 | D & A |  | ?
| C2 | 4 | ? | ? | ? |
| | P06143 | | P | H | E | R | 0 | SIGNALING PROTEIN |
| 35 | 1mih_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | YES |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1mih_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 1udr_1 | D & A |  | YES
| C2 | 4 | NPS | 1 | PROBYES1 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1vlz_1 | D & A |  | YES
| C2 | 2 | NPS | 1 | PROBYES3 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNAL TRANSDUCTION PROTEIN |
| 35 | 1ymu_1 | D & B |  | YES
| C2 | 2 | NPS | 1 | PROBYES3 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 1ymv_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 35 | 2b1j_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 2b1j_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 2iex_1 | A & B | 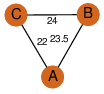 | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | Q5KVX8 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 35 | 2iex_2 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5KVX8 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 35 | 3chy_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNAL TRANSDUCTION PROTEIN |
| 35 | 3f7n_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3f7n_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3fft_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3fft_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3ffw_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3ffw_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3ffx_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3ffx_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3fgz_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3fgz_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvj_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvj_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvk_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvl_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvl_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvm_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvn_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvn_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvo_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvp_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvp_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvq_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvr_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvr_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvs_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 3rvs_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 35 | 5chy_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli K12 | SIGNAL TRANSDUCTION PROTEIN |
| 35 | 6chy_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli K12 | SIGNAL TRANSDUCTION PROTEIN |
| 35 | 6chy_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli K12 | SIGNAL TRANSDUCTION PROTEIN |
| 35 | 6chy_3 | D & A |  | PROBYES
| C2 | 2 | NPS | 1 | PROBYES3 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli K12 | SIGNAL TRANSDUCTION PROTEIN |
| 34 | 1c4w_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 1d4z_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 1dbw_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 34 | 1dbw_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 34 | 1dck_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 34 | 1dck_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 34 | 1dcm_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 34 | 1dcm_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 34 | 1e6k_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | 1E6K | | P | H | E | R | ESCHERICHIA COLI | SIGNALING PROTEIN |
| 34 | 1e6l_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | ESCHERICHIA COLI | SIGNALING PROTEIN |
| 34 | 1e6m_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | ESCHERICHIA COLI K-12 | SIGNALING PROTEIN |
| 34 | 1ehc_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNAL TRANSDUCTION |
| 34 | 1zdm_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 1zdm_2 | D & B |  | PROBNOT
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 2id7_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 2id9_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 2idm_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 2qsj_1 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q5LQW4 | | P | H | E | R | Silicibacter pomeroyi DSS-3 | TRANSCRIPTION |
| 34 | 2qsj_2 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q5LQW4 | | P | H | E | R | Silicibacter pomeroyi DSS-3 | TRANSCRIPTION |
| 34 | 2qsj_3 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q5LQW4 | | P | H | E | R | Silicibacter pomeroyi DSS-3 | TRANSCRIPTION |
| 34 | 3myy_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli K-12 | SIGNALING PROTEIN |
| 34 | 3olv_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3olv_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3olw_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3olw_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3olx_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3olx_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3oly_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3oly_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3oo0_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3oo0_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3oo1_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3oo1_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AE67 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 34 | 3t8y_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYN9 | | P | H | E | R | Thermotoga maritima | HYDROLASE |
| 34 | 3t8y_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYN9 | | P | H | E | R | Thermotoga maritima | HYDROLASE |
| 33 | 1d5w_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | GENE REGULATION |
| 33 | 1d5w_2 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P10958 | | P | H | E | R | Sinorhizobium meliloti | GENE REGULATION |
| 33 | 1hey_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | CHEMOTAXIS |
| 33 | 2che_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNAL TRANSDUCTION PROTEIN |
| 33 | 2chf_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNAL TRANSDUCTION PROTEIN |
| 33 | 2chy_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNAL TRANSDUCTION PROTEIN |
| 33 | 2fka_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fka_2 | D & A |  | PROBYES
| Tetr | 12 | NPS | 1 | NA1 |
Tetr | 12 | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2flk_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2flk_2 | D & o |  | PROBYES
| Octa | 24 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2flw_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmf_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmh_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmh_2 | D & o |  | PROBYES
| Octa | 24 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmh_3 | D & A |  | PROBYES
| C3 | 3 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmi_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmi_2 | D & A |  | PROBYES
| C3 | 3 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2fmk_1 | D & A |  | PROBNOT
| NPS | 1 | NPS | 1 | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2pbp_1 | A & A |  | ?
| C2 | 2 | ? | ? | YES |
D3 | 6 | Q5KYB2 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 33 | 2pbp_2 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5KYB2 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 33 | 2pl9_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTIEN |
| 33 | 2pl9_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTIEN |
| 33 | 2pl9_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTIEN |
| 33 | 2pmc_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2pmc_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2pmc_3 | D & C |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2pmc_4 | D & D |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0A2D5 | | P | H | E | R | Salmonella typhimurium | SIGNALING PROTEIN |
| 33 | 2qq3_1 | A & C |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5KYB2 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 33 | 2qq3_2 | A & F |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5KYB2 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 33 | 3a0u_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9X181 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 33 | 3qyr_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q73VV7 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | LYASE |
| 33 | 3qyr_2 | A & B |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q73VV7 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | LYASE |
| 32 | 1u8t_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 32 | 1u8t_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 32 | 1u8t_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 32 | 1u8t_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 32 | 1u8t_5 | D & C |  | ?
| C2 | 2 | ? | ? | ? |
| | P06143 | | P | H | E | R | Escherichia coli | SIGNALING PROTEIN |
| 32 | 3qxi_1 | A & C | 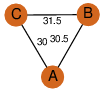 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B2HMZ1 | | P | H | E | R | Mycobacterium marinum | LYASE |
| 32 | 3qxz_1 | A & B | 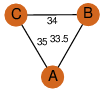 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1MEE0 | | P | H | E | R | Mycobacterium abscessus | LYASE,ISOMERASE |
| 32 | 3r9q_1 | A & C | 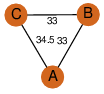 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1MGI6 | | P | H | E | R | Mycobacterium abscessus | LYASE,ISOMERASE |
| 32 | 3t88_1 | A & C |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE/LYASE INHIBITOR |
| 32 | 3t89_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE |
| 32 | 4els_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE |
| 32 | 4elw_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE |
| 32 | 4elx_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE |
| 32 | 4eml_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P73495 | | P | H | E | R | Synechocystis sp. | LYASE |
| 32 | 4i42_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE |
| 32 | 4i42_2 | A & L |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P0ABU0 | | P | H | E | R | Escherichia coli | LYASE |
| 32 | 4i4z_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P73495 | | P | H | E | R | Synechocystis sp. | LYASE |
| 32 | 4i4z_2 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P73495 | | P | H | E | R | Synechocystis sp. | LYASE |
| 32 | 4i52_1 | A & D | 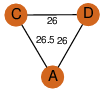 | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | P73495 | | P | H | E | R | 0 | LYASE |
| 32 | 4i52_2 | A & F | 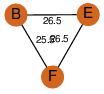 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | P73495 | | P | H | E | R | 0 | LYASE |
| 32 | 4i52_3 | A & I | 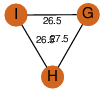 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | P73495 | | P | H | E | R | 0 | LYASE |
| 31 | 1l5y_1 | D & A |  | YES
| C2 | 2 | C2 | 2 | PROBNOT |
C2 | 2 | P13632 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION REGULATOR |
| 31 | 1l5z_1 | D & B |  | NO
| C2 | 2 | C2 | 2 | PROBNOT |
C2 | 2 | P13632 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION REGULATOR |
| 31 | 1l5z_2 | D & B |  | YES
| D2 | 4 | C2 | 2 | NA1 |
D2 | 4 | P13632 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION REGULATOR |
| 31 | 1mvo_1 | D & B |  | PROBNOT
| C2 | 2 | NS | 2 | ? |
| | P13792 | | P | H | E | R | Bacillus subtilis | Transcription |
| 31 | 1qkk_1 | D & B |  | NO
| C2 | 2 | C2 | 2 | PROBNOT |
C2 | 2 | P13632 | | P | H | E | R | SINORHIZOBIUM MELILOTI | TRANSCRIPTIONAL REGULATORY PROTEIN |
| 31 | 3a0r_1 | D & B |  | ?
| C2 | 4 | ? | ? | ? |
| | Q9X180 | | P | H | E | R | Thermotoga maritima | TRANSFERASE |
| 31 | 3a10_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9X181 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 31 | 3grc_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q121U0 | | P | H | E | R | Polaromonas sp. JS666 | TRANSFERASE |
| 31 | 3grc_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q121U0 | | P | H | E | R | Polaromonas sp. JS666 | TRANSFERASE |
| 31 | 3grc_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q121U0 | | P | H | E | R | Polaromonas sp. JS666 | TRANSFERASE |
| 31 | 3grc_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q121U0 | | P | H | E | R | Polaromonas sp. JS666 | TRANSFERASE |
| 31 | 3grc_5 | D & A |  | ?
| NS | 4 | ? | ? | ? |
| | Q121U0 | | P | H | E | R | Polaromonas sp. JS666 | TRANSFERASE |
| 31 | 3h02_1 | A & D |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q7CQ56 | | P | H | E | R | Salmonella typhimurium | LYASE |
| 31 | 3h02_2 | A & A | 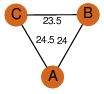 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q7CQ56 | | P | H | E | R | Salmonella typhimurium | LYASE |
| 31 | 3h02_3 | A & E | 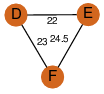 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q7CQ56 | | P | H | E | R | Salmonella typhimurium | LYASE |
| 31 | 3h1g_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P71403 | | P | H | E | R | Helicobacter pylori | SIGNALING PROTEIN |
| 31 | 3nhz_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | C6DXJ2 | | P | H | E | R | Mycobacterium tuberculosis | DNA BINDING PROTEIN |
| 31 | 3nhz_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | C6DXJ2 | | P | H | E | R | Mycobacterium tuberculosis | DNA BINDING PROTEIN |
| 31 | 3nhz_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | C6DXJ2 | | P | H | E | R | Mycobacterium tuberculosis | DNA BINDING PROTEIN |
| 31 | 3nhz_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | C6DXJ2 | | P | H | E | R | Mycobacterium tuberculosis | DNA BINDING PROTEIN |
| 31 | 3nnn_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9WYN0 | | P | H | E | R | Thermotoga maritima | DNA BINDING PROTEIN |
| 31 | 3qka_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | B2HSK3 | | P | H | E | R | Mycobacterium marinum M | LYASE |
| 31 | 4hnq_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9KQD5 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 1tmy_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | CHEMOTAXIS |
| 30 | 1u0s_1 | D & Y |  | ?
| NPS | 2 | ? | ? | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 30 | 1zh2_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P21866 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 30 | 1zh4_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P21866 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 30 | 2tmy_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | CHEMOTAXIS |
| 30 | 3b2n_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | Q99UF4 | | P | H | E | R | Staphylococcus aureus | TRANSCRIPTION |
| 30 | 3eod_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P0AEV1 | | P | H | E | R | Escherichia coli K12 | SIGNALING PROTEIN |
| 30 | 3gwg_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P71403 | | P | H | E | R | Helicobacter pylori | SIGNALING PROTEIN |
| 30 | 3h1e_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P71403 | | P | H | E | R | Helicobacter pylori | SIGNALING PROTEIN |
| 30 | 3mm4_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | O22267 | | P | H | E | R | Arabidopsis thaliana | TRANSFERASE |
| 30 | 3mmn_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | O22267 | | P | H | E | R | Arabidopsis thaliana | TRANSFERASE |
| 30 | 3r0o_1 | A & A | 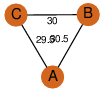 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QFV0 | | P | H | E | R | Mycobacterium avium | LYASE |
| 30 | 3tmy_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | CHEMOTAXIS |
| 30 | 3tmy_2 | D & B |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | CHEMOTAXIS |
| 30 | 3to5_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9KQD5 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 4h60_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | C3M734 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 4hnr_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9KKK8 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 4hns_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9KQD5 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 4iga_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN/MOTOR PROTEIN |
| 30 | 4jp1_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | A5F6J9 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 4l85_1 | D & B |  | ?
| NS | 2 | ? | ? | YES |
C2 | 2 | P21866 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 30 | 4l85_2 | D & C |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | P21866 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 30 | 4lx8_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | A5F6J9 | | P | H | E | R | Vibrio cholerae | SIGNALING PROTEIN |
| 30 | 4tmy_1 | D & B |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | CHEMOTAXIS |
| 30 | 4tmy_2 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q56312 | | P | H | E | R | Thermotoga maritima | CHEMOTAXIS |
| 29 | 1xx4_1 | A & C | 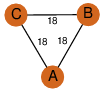 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P23965 | | P | H | E | R | Rattus norvegicus | isomerase |
| 29 | 1xx4_2 | A & A |  | ?
| C2 | 2 | ? | ? | ? |
| | P23965 | | P | H | E | R | Rattus norvegicus | isomerase |
| 29 | 2pkx_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P23836 | | P | H | E | R | Escherichia coli | TRANSCRIPTIONAL REGULATOR |
| 29 | 2pl1_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P23836 | | P | H | E | R | Escherichia coli | TRANSCRIPTIONAL REGULATOR |
| 29 | 3f6c_1 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | P0ACZ4 | | P | H | E | R | Escherichia coli K-12 | DNA BINDING PROTEIN |
| 29 | 3f6c_2 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P0ACZ4 | | P | H | E | R | Escherichia coli K-12 | DNA BINDING PROTEIN |
| 29 | 3f6c_3 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | P0ACZ4 | | P | H | E | R | Escherichia coli K-12 | DNA BINDING PROTEIN |
| 29 | 3h1f_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | P71403 | | P | H | E | R | Helicobacter pylori | SIGNALING PROTEIN |
| 29 | 3hdv_1 | D & B |  | ?
| C2 | 4 | ? | ? | PROBYES1 |
C2 | 2 | Q88QX9 | | P | H | E | R | Pseudomonas putida | transcription regulator |
| 29 | 3hdv_2 | D & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q88QX9 | | P | H | E | R | Pseudomonas putida | transcription regulator |
| 29 | 3hdv_3 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q88QX9 | | P | H | E | R | Pseudomonas putida | transcription regulator |
| 29 | 3i42_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1H0I3 | | P | H | E | R | Methylobacillus flagellatus KT | Structural Genomics, Unknown function |
| 29 | 3lke_1 | A & B | 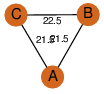 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9KBS5 | | P | H | E | R | Bacillus halodurans | LYASE |
| 29 | 3t6k_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | A9WCL5 | | P | H | E | R | Chloroflexus aurantiacus | SIGNALING PROTEIN |
| 29 | 3t6k_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | A9WCL5 | | P | H | E | R | Chloroflexus aurantiacus | SIGNALING PROTEIN |
| 29 | 3trr_1 | A & A | 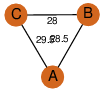 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1ML12 | | P | H | E | R | Mycobacterium abscessus | ISOMERASE |
| 29 | 3trr_2 | A & F | 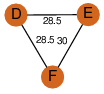 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1ML12 | | P | H | E | R | Mycobacterium abscessus | ISOMERASE |
| 28 | 1dcf_1 | D & A |  | NO
| C2 | 2 | C2 | 2 | ? |
| | P49333 | | P | H | E | R | Arabidopsis thaliana | TRANSFERASE |
| 28 | 1oxb_1 | D & B |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_1 | D & B |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_2 | D & D |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_3 | D & F |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_4 | D & H |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_5 | D & J |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_6 | D & L |  | PROBNOT
| NPS | 2 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 1oxk_7 | D & F |  | PROBYES
| NS | 8 | NPS | 2 | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | SIGNALING PROTEIN |
| 28 | 2r25_1 | D & B |  | ?
| NPS | 2 | ? | ? | ? |
| | Q07688 | | P | H | E | R | Saccharomyces cerevisiae | Signaling PROTEIN/Transferase |
| 28 | 2uzf_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5HH38 | | P | H | E | R | STAPHYLOCOCCUS AUREUS | LYASE |
| 28 | 2w3p_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q13WK7 | | P | H | E | R | BURKHOLDERIA XENOVORANS | LYASE |
| 28 | 3h81_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3h81_2 | A & B | 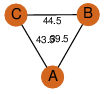 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3jte_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | A3DCZ0 | | P | H | E | R | Clostridium thermocellum ATCC 27405 | PROTEIN BINDING |
| 28 | 3pe8_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | A0QVP0 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 28 | 3pzk_1 | A & A |  | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3pzk_2 | A & F |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3q0g_1 | A & A |  | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3q0g_2 | A & F |  | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3q0g_3 | A & D |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3q0g_4 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 28 | 3q0j_1 | A & A |  | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE/LYASE INHIBITOR |
| 28 | 3q0j_2 | A & F |  | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE/LYASE INHIBITOR |
| 28 | 3q0j_3 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE/LYASE INHIBITOR |
| 28 | 3q0j_4 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE/LYASE INHIBITOR |
| 28 | 3rsi_1 | A & B | 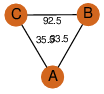 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1MIA8 | | P | H | E | R | Mycobacterium abscessus | ISOMERASE |
| 28 | 4f47_1 | A & A | 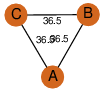 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B2HI06 | | P | H | E | R | Mycobacterium marinum | LYASE |
| 28 | 4fzw_1 | A & A |  | ?
| D3 | 12 | ? | ? | ? |
| | P76082 | | P | H | E | R | Escherichia coli | ISOMERASE/LYASE |
| 28 | 4qfe_1 | A & A | 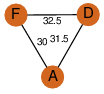 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QS88 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 28 | 4qfe_2 | A & H | 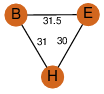 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QS88 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 28 | 4qfe_3 | A & K | 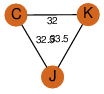 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QS88 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 28 | 4qfe_4 | A & L | 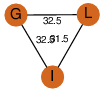 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QS88 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 27 | 1b00_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P08402 | | P | H | E | R | Escherichia coli | GENE REGULATION |
| 27 | 1b00_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P08402 | | P | H | E | R | Escherichia coli | GENE REGULATION |
| 27 | 1dub_1 | A & A |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | P14604 | | P | H | E | R | 0 | LYASE |
| 27 | 1dz3_1 | D & B |  | NO
| C2 | 2 | C2 | 2 | ? |
| | CAA05307 | | P | H | E | R | BACILLUS STEAROTHERMOPHILUS | RESPONSE REGULATOR |
| 27 | 1ey3_1 | A & E |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | P14604 | | P | H | E | R | Rattus norvegicus | LYASE |
| 27 | 1mj3_1 | A & A |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | P14604 | | P | H | E | R | Rattus norvegicus | LYASE |
| 27 | 1zes_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P08402 | | P | H | E | R | Escherichia coli | TRANSCRIPTION ACTIVATOR |
| 27 | 1zes_2 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P08402 | | P | H | E | R | Escherichia coli | TRANSCRIPTION ACTIVATOR |
| 27 | 2dub_1 | A & E |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | P14604 | | P | H | E | R | 0 | LYASE |
| 27 | 2dub_2 | A & E |  | YES
| C2 | 6 | D3 | 6 | PROBYES3 |
D3 | 6 | P14604 | | P | H | E | R | 0 | LYASE |
| 27 | 2iyn_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 27 | 2iyn_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 27 | 2iyn_3 | D & C |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 27 | 2iyn_4 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 27 | 2iyn_5 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 27 | 2jk1_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P26408 | | P | H | E | R | RHODOBACTER CAPSULATUS | DNA-BINDING |
| 27 | 2qr3_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q64UD2 | | P | H | E | R | Bacteroides fragilis | TRANSCRIPTION |
| 27 | 2vui_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P26408 | | P | H | E | R | RHODOBACTER CAPSULATUS | DNA-BINDING |
| 27 | 3eul_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | O53856 | | P | H | E | R | Mycobacterium tuberculosis | TRANSCRIPTION |
| 27 | 3eul_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | O53856 | | P | H | E | R | Mycobacterium tuberculosis | TRANSCRIPTION |
| 27 | 3eul_3 | D & C |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | O53856 | | P | H | E | R | Mycobacterium tuberculosis | TRANSCRIPTION |
| 27 | 3eul_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | O53856 | | P | H | E | R | Mycobacterium tuberculosis | TRANSCRIPTION |
| 27 | 3eul_5 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | O53856 | | P | H | E | R | Mycobacterium tuberculosis | TRANSCRIPTION |
| 27 | 3eul_6 | D & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O53856 | | P | H | E | R | Mycobacterium tuberculosis | TRANSCRIPTION |
| 27 | 3hin_1 | A & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q6N8W7 | | P | H | E | R | Rhodopseudomonas palustris | LYASE |
| 27 | 3hin_2 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q6N8W7 | | P | H | E | R | Rhodopseudomonas palustris | LYASE |
| 27 | 3hin_3 | A & B | 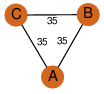 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q6N8W7 | | P | H | E | R | Rhodopseudomonas palustris | LYASE |
| 27 | 3hin_4 | A & B | 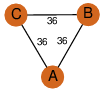 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q6N8W7 | | P | H | E | R | Rhodopseudomonas palustris | LYASE |
| 27 | 3kqf_1 | A & A | 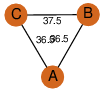 | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | Q81Q82 | | P | H | E | R | Bacillus anthracis | ISOMERASE |
| 27 | 3kqf_2 | A & F | 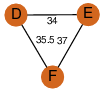 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q81Q82 | | P | H | E | R | Bacillus anthracis | ISOMERASE |
| 27 | 3moy_1 | A & C | 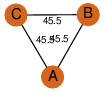 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0R747 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 27 | 4e7o_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q8DNC2 | | P | H | E | R | Streptococcus pneumoniae | TRANSCRIPTION REGULATOR |
| 27 | 4e7p_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q8DNC2 | | P | H | E | R | Streptococcus pneumoniae | TRANSCRIPTION REGULATOR |
| 27 | 4e7p_2 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q8DNC2 | | P | H | E | R | Streptococcus pneumoniae | TRANSCRIPTION REGULATOR |
| 27 | 4fjw_1 | A & F | 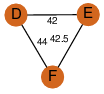 | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 27 | 4fjw_2 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P64016 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 27 | 4nic_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | G0GNT0 | | P | H | E | R | Klebsiella pneumoniae | TRANSCRIPTION REGULATOR |
| 27 | 4nic_2 | D & D |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | G0GNT0 | | P | H | E | R | Klebsiella pneumoniae | TRANSCRIPTION REGULATOR |
| 26 | 1f51_1 | D & F |  | ?
| C2 | 4 | ? | ? | ? |
| | P06535 | | P | H | E | R | Bacillus subtilis | TRANSFERASE |
| 26 | 1f51_2 | D & G |  | ?
| C2 | 4 | ? | ? | ? |
| | P06535 | | P | H | E | R | Bacillus subtilis | TRANSFERASE |
| 26 | 1hzd_1 | A & A |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | Q13825 | | P | H | E | R | Homo sapiens | LYASE |
| 26 | 1nat_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | PROBYES2 |
C2 | 2 | P06628 | | P | H | E | R | Bacillus subtilis | REGULATORY PROTEIN |
| 26 | 1nxo_1 | D & B |  | NO
| C2 | 2 | C2 | 2 | PROBYES3 |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 1nxp_1 | D & A |  | NO
| C2 | 2 | C2 | 2 | YES |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 1nxs_1 | D & B |  | NO
| C2 | 2 | C2 | 2 | PROBNOT |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 1nxv_1 | D & A |  | NO
| C2 | 2 | C2 | 2 | PROBYES3 |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 1nxw_1 | D & A |  | NO
| C2 | 2 | C2 | 2 | PROBNOT |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 1nxx_1 | D & A |  | NO
| C2 | 2 | C2 | 2 | PROBYES3 |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 1pey_1 | D & A |  | PROBYES
| C3 | 3 | NPS | 1 | ? |
| | P06628 | | P | H | E | R | Bacillus subtilis | TRANSFERASE |
| 26 | 1qmp_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | CAA05307 | | P | H | E | R | BACILLUS STEAROTHERMOPHILUS | REPLICATION |
| 26 | 1qmp_2 | D & B |  | NO
| NPS | 1 | NPS | 1 | ? |
| | CAA05307 | | P | H | E | R | BACILLUS STEAROTHERMOPHILUS | REPLICATION |
| 26 | 1qmp_3 | D & C |  | NO
| NPS | 1 | NPS | 1 | ? |
| | CAA05307 | | P | H | E | R | BACILLUS STEAROTHERMOPHILUS | REPLICATION |
| 26 | 1qmp_4 | D & D |  | NO
| NPS | 1 | NPS | 1 | ? |
| | CAA05307 | | P | H | E | R | BACILLUS STEAROTHERMOPHILUS | REPLICATION |
| 26 | 1srr_1 | D & A |  | PROBYES
| C2 | 2 | NPS | 1 | PROBNOT |
C2 | 2 | P06628 | | P | H | E | R | Bacillus subtilis | REGULATORY PROTEIN |
| 26 | 1srr_2 | D & C |  | PROBYES
| C2 | 2 | NPS | 1 | PROBYES3 |
C2 | 2 | P06628 | | P | H | E | R | Bacillus subtilis | REGULATORY PROTEIN |
| 26 | 1zy2_1 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | O67198 | | P | H | E | R | Aquifex aeolicus | TRANSCRIPTION |
| 26 | 2a9o_1 | D & A |  | PROBYES
| NPS | 1 | C2 | 2 | YES |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 2a9p_1 | D & A |  | PROBYES
| NPS | 1 | C2 | 2 | PROBYES2 |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 2a9q_1 | D & A |  | PROBYES
| NPS | 1 | C2 | 2 | YES |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 26 | 2ftk_1 | D & F |  | ?
| C2 | 4 | ? | ? | ? |
| | P06535 | | P | H | E | R | Bacillus subtilis | TRANSFERASE |
| 26 | 2ftk_2 | D & G |  | ?
| C2 | 4 | ? | ? | ? |
| | P06535 | | P | H | E | R | Bacillus subtilis | TRANSFERASE |
| 26 | 2gtr_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9Y232 | | P | H | E | R | Homo sapiens | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 27 | 2jba_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 26 | 2qxy_1 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q9WZU6 | | P | H | E | R | Thermotoga maritima | TRANSCRIPTION |
| 26 | 2qxy_2 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | Q9WZU6 | | P | H | E | R | Thermotoga maritima | TRANSCRIPTION |
| 26 | 2qxy_3 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | Q9WZU6 | | P | H | E | R | Thermotoga maritima | TRANSCRIPTION |
| 26 | 2vuh_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P26408 | | P | H | E | R | RHODOBACTER CAPSULATUS | DNA-BINDING |
| 26 | 2zqq_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q13825 | | P | H | E | R | Homo sapiens | LYASE |
| 26 | 2zqr_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q13825 | | P | H | E | R | Homo sapiens | LYASE |
| 26 | 3cg0_1 | D & A |  | ?
| C2 | 4 | ? | ? | ? |
| | Q311G8 | | P | H | E | R | Desulfovibrio desulfuricans subsp. desulfuricans str. | LYASE |
| 26 | 3cg0_2 | D & A |  | ?
| C2 | 4 | ? | ? | ? |
| | Q311G8 | | P | H | E | R | Desulfovibrio desulfuricans subsp. desulfuricans str. | LYASE |
| 26 | 3dge_1 | D & C |  | ?
| NPS | 4 | ? | ? | ? |
| | Q9WZV7 | | P | H | E | R | Thermotoga maritima | TRANSFERASE/SIGNALING PROTEIN |
| 26 | 3dge_2 | D & D |  | ?
| NPS | 2 | ? | ? | ? |
| | Q9WZV7 | | P | H | E | R | Thermotoga maritima | TRANSFERASE/SIGNALING PROTEIN |
| 26 | 3dge_3 | D & C |  | ?
| NPS | 2 | ? | ? | ? |
| | Q9WZV7 | | P | H | E | R | Thermotoga maritima | TRANSFERASE/SIGNALING PROTEIN |
| 26 | 3dgf_1 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYT9 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 26 | 3hrx_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5SLK3 | | P | H | E | R | Thermus thermophilus | LYASE |
| 26 | 3hrx_2 | A & F |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5SLK3 | | P | H | E | R | Thermus thermophilus | LYASE |
| 26 | 3hv2_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q4K707 | | P | H | E | R | Pseudomonas fluorescens Pf-5 | SIGNALING PROTEIN |
| 26 | 3kht_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | Q2SI73 | | P | H | E | R | Hahella chejuensis | SIGNALING PROTEIN |
| 26 | 3kht_2 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q2SI73 | | P | H | E | R | Hahella chejuensis | SIGNALING PROTEIN |
| 26 | 3pea_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q81L70 | | P | H | E | R | Bacillus anthracis | ISOMERASE |
| 26 | 3pea_2 | A & E |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q81L70 | | P | H | E | R | Bacillus anthracis | ISOMERASE |
| 26 | 3q15_1 | D & D |  | ?
| C2 | 4 | ? | ? | ? |
| | Q59HN8 | | P | H | E | R | Bacillus subtilis | HYDROLASE/KINASE |
| 26 | 3q15_2 | D & D |  | ?
| C2 | 4 | ? | ? | ? |
| | Q59HN8 | | P | H | E | R | Bacillus subtilis | HYDROLASE/KINASE |
| 26 | 3r9s_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QC84 | | P | H | E | R | Mycobacterium avium | LYASE |
| 26 | 3r9t_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q73WL5 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | LYASE |
| 26 | 3rrv_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q743A2 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | ISOMERASE |
| 26 | 3w9s_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | C4X5T7 | | P | H | E | R | Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | SIGNALING PROTEIN/ANTIMICROBIAL PROTEIN |
| 26 | 3w9s_2 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | C4X5T7 | | P | H | E | R | Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | SIGNALING PROTEIN/ANTIMICROBIAL PROTEIN |
| 26 | 3w9s_3 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | C4X5T7 | | P | H | E | R | Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | SIGNALING PROTEIN/ANTIMICROBIAL PROTEIN |
| 26 | 4di1_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B2HHI0 | | P | H | E | R | Mycobacterium marinum | Lyase |
| 26 | 4izb_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5LLW6 | | P | H | E | R | Ruegeria pomeroyi | HYDROLASE |
| 26 | 4jjt_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | I6YEH6 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 26 | 4q7e_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | B0SGG3 | | P | H | E | R | Leptospira biflexa serovar Patoc | SIGNALING PROTEIN |
| 25 | 1nxt_1 | D & A |  | NO
| C2 | 2 | C2 | 2 | PROBYES3 |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 25 | 1uiy_1 | A & A |  | PROBNOT
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | P83702 | | P | H | E | R | Thermus thermophilus | Lyase |
| 25 | 2a9r_1 | D & A |  | ?
| C2 | 2 | ? | ? | YES |
C2 | 2 | Q9S1K0 | | P | H | E | R | Streptococcus pneumoniae | SIGNALING PROTEIN |
| 25 | 2hw5_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P30084 | | P | H | E | R | Homo sapiens | LYASE |
| 25 | 2jb9_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 25 | 2jb9_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P0AFJ5 | | P | H | E | R | ESCHERICHIA COLI | TRANSCRIPTION |
| 25 | 2np9_1 | A & A |  | ?
| D3 | 6 | ? | ? | NA1 |
D3 | 6 | Q8KLK7 | | P | H | E | R | Streptomyces toyocaensis | OXIDOREDUCTASE |
| 25 | 2np9_2 | A & A | 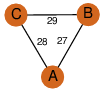 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q8KLK7 | | P | H | E | R | Streptomyces toyocaensis | OXIDOREDUCTASE |
| 25 | 2qvg_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q5ZSR0 | | P | H | E | R | Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | TRANSFERASE |
| 25 | 2rjn_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q2BKU2 | | P | H | E | R | Neptuniibacter caesariensis | HYDROLASE |
| 25 | 2zay_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | Q1JZD9 | | P | H | E | R | Desulfuromonas acetoxidans | SIGNALING PROTEIN |
| 25 | 2zay_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1JZD9 | | P | H | E | R | Desulfuromonas acetoxidans | SIGNALING PROTEIN |
| 25 | 3gkb_1 | A & B | 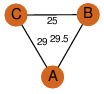 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q82QL3 | | P | H | E | R | Streptomyces avermitilis | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 25 | 3gl9_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYT9 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 25 | 3gl9_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYT9 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 25 | 3gl9_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYT9 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 25 | 3gl9_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYT9 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 25 | 3kyi_1 | D & B |  | ?
| NPS | 2 | ? | ? | ? |
| | Q8KLS0 | | P | H | E | R | Rhodobacter sphaeroides | TRANSFERASE |
| 25 | 3kyj_1 | D & B |  | ?
| NPS | 2 | ? | ? | ? |
| | Q8KLS0 | | P | H | E | R | Rhodobacter sphaeroides | TRANSFERASE |
| 25 | 3lao_1 | A & A | 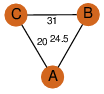 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9I498 | | P | H | E | R | Pseudomonas aeruginosa | lyase, isomerase |
| 25 | 3oc7_1 | A & B | 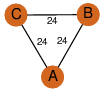 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QBQ2 | | P | H | E | R | Mycobacterium avium | Lyase |
| 25 | 3p85_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | A0QIX8 | | P | H | E | R | Mycobacterium avium | LYASE |
| 25 | 4izc_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5LLW6 | | P | H | E | R | Ruegeria pomeroyi | hydrolase/substrate |
| 25 | 4izd_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5LLW6 | | P | H | E | R | Ruegeria pomeroyi | hydrolase/substrate |
| 25 | 4ja2_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9WYT9 | | P | H | E | R | Thermotoga maritima | SIGNALING PROTEIN |
| 25 | 4jas_1 | D & D |  | ?
| C2 | 4 | ? | ? | ? |
| | Q9WZV7 | | P | H | E | R | Thermotoga maritima | TRANSFERASE/SIGNALING PROTEIN |
| 25 | 4jav_1 | D & C |  | ?
| C2 | 4 | ? | ? | ? |
| | Q9WZV7 | | P | H | E | R | Thermotoga maritima | TRANSFERASE/SIGNALING PROTEIN |
| 25 | 4nnq_1 | A & A | 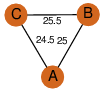 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q8GGP6 | | P | H | E | R | Streptomyces atroolivaceus | LYASE |
| 24 | 1k66_1 | D & B |  | NO
| C2 | 2 | C2 | 2 | PROBNOT |
C2 | 2 | Q8RTM8 | | P | H | E | R | Tolypothrix sp. PCC 7601 | SIGNALING PROTEIN |
| 24 | 1m5t_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q9A5I4 | | P | H | E | R | Caulobacter vibrioides | SIGNALING PROTEIN, CELL CYCLE |
| 24 | 1m5u_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q9A5I4 | | P | H | E | R | Caulobacter vibrioides | SIGNALING PROTEIN, CELL CYCLE |
| 24 | 1mav_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q9A5I4 | | P | H | E | R | Caulobacter vibrioides | SIGNALING PROTEIN, CELL CYCLE |
| 24 | 1mb0_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q9A5I4 | | P | H | E | R | Caulobacter vibrioides | SIGNALING PROTEIN, CELL CYCLE |
| 24 | 1mb3_1 | D & A |  | NO
| NPS | 1 | NPS | 1 | ? |
| | Q9A5I4 | | P | H | E | R | Caulobacter vibrioides | SIGNALING PROTEIN, CELL CYCLE |
| 24 | 1q51_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 24 | 1q51_2 | A & L |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 24 | 1q52_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 24 | 1q52_2 | A & K |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 24 | 1sg4_1 | A & A | 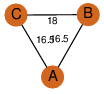 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P42126 | | P | H | E | R | Homo sapiens | ISOMERASE |
| 24 | 1sg4_2 | A & A |  | ?
| C2 | 6 | ? | ? | PROBYES1 |
C3 | 3 | P42126 | | P | H | E | R | Homo sapiens | ISOMERASE |
| 24 | 1xhf_1 | D & B |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | P0A9Q1 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 24 | 2ppy_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5KYF9 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 24 | 2ppy_2 | A & A | 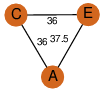 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q5KYF9 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 24 | 2ppy_3 | A & F | 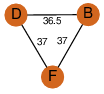 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q5KYF9 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 24 | 2zwm_1 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | P37478 | | P | H | E | R | Bacillus subtilis | Transcription |
| 24 | 3c3m_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | A3CS79 | | P | H | E | R | Methanoculleus marisnigri JR1 | SIGNALING PROTEIN |
| 24 | 3cz5_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1YEP2 | | P | H | E | R | Aurantimonas sp. | TRANSCRIPTION REGULATOR |
| 24 | 3cz5_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1YEP2 | | P | H | E | R | Aurantimonas sp. | TRANSCRIPTION REGULATOR |
| 24 | 3cz5_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1YEP2 | | P | H | E | R | Aurantimonas sp. | TRANSCRIPTION REGULATOR |
| 24 | 3cz5_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1YEP2 | | P | H | E | R | Aurantimonas sp. | TRANSCRIPTION REGULATOR |
| 24 | 3f6p_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | P37478 | | P | H | E | R | Bacillus subtilis | TRANSCRIPTION REGULATOR |
| 24 | 3gt7_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q2LQE8 | | P | H | E | R | Syntrophus aciditrophicus SB | HYDROLASE |
| 24 | 3lte_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_10 | D & S |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_11 | D & U |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_12 | D & W |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_2 | D & D |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_3 | D & E |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_4 | D & H |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_5 | D & J |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_6 | D & K |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_7 | D & N |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_8 | D & O |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3lte_9 | D & Q |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q1N036 | | P | H | E | R | Bermanella marisrubri | TRANSCRIPTION |
| 24 | 3swx_1 | A & B | 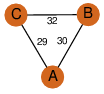 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1MM02 | | P | H | E | R | Mycobacterium abscessus | ISOMERASE |
| 24 | 3t3w_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | G7CL68 | | P | H | E | R | Mycobacterium thermoresistibile | LYASE |
| 24 | 4jfc_1 | A & A | 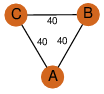 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q129C0 | | P | H | E | R | Polaromonas sp. JS666 | LYASE |
| 24 | 4knp_1 | A & A | 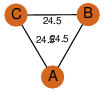 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q742C8 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | LYASE |
| 24 | 4mou_1 | A & C | 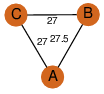 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A1RAA6 | | P | H | E | R | Arthrobacter aurescens | ISOMERASE |
| 23 | 1xhe_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0A9Q1 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 23 | 2fw2_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q9Y6F7 | | P | H | E | R | Homo sapiens | GENE REGULATION |
| 23 | 2fw2_2 | A & E | 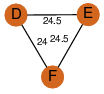 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q9Y6F7 | | P | H | E | R | Homo sapiens | GENE REGULATION |
| 23 | 2fw2_3 | A & B | 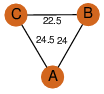 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q9Y6F7 | | P | H | E | R | Homo sapiens | GENE REGULATION |
| 23 | 2q35_1 | A & A | 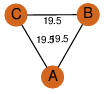 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q6DNE7 | | P | H | E | R | Lyngbya majuscula | LYASE |
| 23 | 3c97_1 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q2U940 | | P | H | E | R | Aspergillus oryzae RIB40 | SIGNALING PROTEIN, TRANSFERASE |
| 23 | 3g64_1 | A & A | 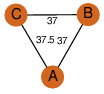 | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | Q93JE8 | | P | H | E | R | Streptomyces coelicolor A3(2) | LYASE |
| 23 | 3g64_2 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q93JE8 | | P | H | E | R | Streptomyces coelicolor A3(2) | LYASE |
| 23 | 3hdg_1 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q7MRH8 | | P | H | E | R | Wolinella succinogenes | structural genomics, unknown function |
| 23 | 3hdg_2 | D & D |  | ?
| C2 | 2 | ? | ? | ? |
| | Q7MRH8 | | P | H | E | R | Wolinella succinogenes | structural genomics, unknown function |
| 23 | 3hdg_3 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q7MRH8 | | P | H | E | R | Wolinella succinogenes | structural genomics, unknown function |
| 23 | 3hdg_4 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q7MRH8 | | P | H | E | R | Wolinella succinogenes | structural genomics, unknown function |
| 23 | 3hzh_1 | D & A |  | ?
| NPS | 2 | ? | ? | ? |
| | O51615 | | P | H | E | R | Borrelia burgdorferi | SIGNALING PROTEIN |
| 23 | 3kto_1 | D & B |  | ?
| C3 | 3 | ? | ? | ? |
| | Q15U25 | | P | H | E | R | Pseudoalteromonas atlantica T6c | transcription regulator |
| 23 | 3kto_2 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q15U25 | | P | H | E | R | Pseudoalteromonas atlantica T6c | transcription regulator |
| 23 | 3kto_3 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q15U25 | | P | H | E | R | Pseudoalteromonas atlantica T6c | transcription regulator |
| 23 | 3kto_4 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q15U25 | | P | H | E | R | Pseudoalteromonas atlantica T6c | transcription regulator |
| 23 | 3n53_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q3A6W4 | | P | H | E | R | Pelobacter carbinolicus | TRANSCRIPTION |
| 23 | 3n53_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q3A6W4 | | P | H | E | R | Pelobacter carbinolicus | TRANSCRIPTION |
| 23 | 3n53_3 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q3A6W4 | | P | H | E | R | Pelobacter carbinolicus | TRANSCRIPTION |
| 23 | 3nhm_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1CZZ7 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 23 | 3nhm_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1CZZ7 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 23 | 3nhm_3 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q1CZZ7 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 23 | 3p5m_1 | A & E |  | ?
| D3 | 6 | ? | ? | ? |
| | A0QFV4 | | P | H | E | R | Mycobacterium avium | ISOMERASE |
| 23 | 4jwv_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q2G8B7 | | P | H | E | R | Novosphingobium aromaticivorans | LYASE |
| 23 | 4nek_1 | A & A |  | ?
| D3 | 6 | ? | ? | YES |
C3 | 3 | Q2VZN8 | | P | H | E | R | Magnetospirillum magneticum | ISOMERASE |
| 23 | 4nek_2 | A & B |  | ?
| C3 | 3 | ? | ? | PROBNOT |
C3 | 3 | Q2VZN8 | | P | H | E | R | Magnetospirillum magneticum | ISOMERASE |
| 23 | 4nek_3 | A & C |  | ?
| C3 | 3 | ? | ? | PROBNOT |
C3 | 3 | Q2VZN8 | | P | H | E | R | Magnetospirillum magneticum | ISOMERASE |
| 22 | 2ej5_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5KYB3 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 22 | 2ej5_2 | A & C |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5KYB3 | | P | H | E | R | Geobacillus kaustophilus | LYASE |
| 22 | 2vx2_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q96DC8 | | P | H | E | R | HOMO SAPIENS | ISOMERASE |
| 22 | 2vx2_2 | A & F |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q96DC8 | | P | H | E | R | HOMO SAPIENS | ISOMERASE |
| 22 | 2vx2_3 | A & G |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q96DC8 | | P | H | E | R | HOMO SAPIENS | ISOMERASE |
| 22 | 3i47_1 | A & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C3 | 3 | Q5ZUH0 | | P | H | E | R | Legionella pneumophila subsp. pneumophila | LYASE |
| 22 | 3i47_2 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5ZUH0 | | P | H | E | R | Legionella pneumophila subsp. pneumophila | LYASE |
| 22 | 3nns_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9WXY0 | | P | H | E | R | Thermotoga maritima | DNA BINDING PROTEIN |
| 22 | 3nns_2 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | Q9WXY0 | | P | H | E | R | Thermotoga maritima | DNA BINDING PROTEIN |
| 22 | 3nns_3 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | Q9WXY0 | | P | H | E | R | Thermotoga maritima | DNA BINDING PROTEIN |
| 21 | 1ef8_1 | A & A | 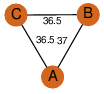 | YES
| C3 | 3 | D3 | 6 | YES |
D3 | 6 | P52045 | | P | H | E | R | 0 | LYASE |
| 21 | 1ef8_2 | A & A |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | P52045 | | P | H | E | R | 0 | LYASE |
| 21 | 1ef9_1 | A & A |  | YES
| NPS | 1 | D3 | 6 | PROBYES2 |
D3 | 6 | P52045 | | P | H | E | R | 0 | LYASE |
| 21 | 2j5i_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O69762 | | P | H | E | R | PSEUDOMONAS FLUORESCENS | LYASE |
| 24 | 2j5i_2 | A & L |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O69762 | | P | H | E | R | PSEUDOMONAS FLUORESCENS | LYASE |
| 21 | 2vss_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O69762 | | P | H | E | R | PSEUDOMONAS FLUORESCENS | LYASE |
| 24 | 2vsu_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O69762 | | P | H | E | R | PSEUDOMONAS FLUORESCENS | LYASE |
| 21 | 3cfy_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q87PN2 | | P | H | E | R | Vibrio parahaemolyticus RIMD 2210633 | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 21 | 3cg4_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q2FQ04 | | P | H | E | R | Methanospirillum hungatei JF-1 | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 21 | 3crn_1 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q2FNF5 | | P | H | E | R | Methanospirillum hungatei JF-1 | SIGNALING PROTEIN |
| 21 | 3h5i_1 | D & A |  | ?
| C3 | 3 | ? | ? | ? |
| | Q3ADQ4 | | P | H | E | R | Carboxydothermus hydrogenoformans Z-2901 | TRANSCRIPTION |
| 21 | 3heb_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q2RWM6 | | P | H | E | R | Rhodospirillum rubrum | transcription regulator |
| 20 | 1k68_1 | D & B |  | YES
| NS | 2 | C2 | 2 | YES |
C2 | 2 | Q8RTN0 | | P | H | E | R | Tolypothrix sp. PCC 7601 | SIGNALING PROTEIN |
| 20 | 3cu5_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | A9KIW7 | | P | H | E | R | Clostridium phytofermentans ISDg | TRANSCRIPTION REGULATOR |
| 20 | 3cu5_2 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | A9KIW7 | | P | H | E | R | Clostridium phytofermentans ISDg | TRANSCRIPTION REGULATOR |
| 20 | 3cu5_3 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | A9KIW7 | | P | H | E | R | Clostridium phytofermentans ISDg | TRANSCRIPTION REGULATOR |
| 19 | 1zgz_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P38684 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 19 | 1zgz_2 | D & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P38684 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 19 | 1zgz_3 | D & A |  | ?
| C2 | 4 | ? | ? | PROBYES1 |
C2 | 2 | P38684 | | P | H | E | R | Escherichia coli | TRANSCRIPTION |
| 0 | 1dci_1 | A & A |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | Q62651 | | P | H | E | R | Rattus norvegicus | LYASE |
| 0 | 1hno_1 | A & E | 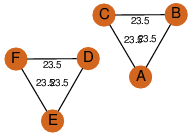 | PROBNOT
| D3 | 6 | D3 | 6 | ? |
| | Q05871 | | P | H | E | R | Saccharomyces cerevisiae | ISOMERASE |
| 0 | 1hno_2 | A & A | 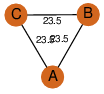 | PROBYES
| C3 | 3 | D3 | 6 | NO |
C3 | 3 | Q05871 | | P | H | E | R | Saccharomyces cerevisiae | ISOMERASE |
| 0 | 1hnu_1 | A & A | 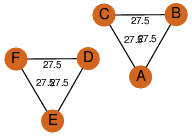 | PROBNOT
| D3 | 6 | D3 | 6 | ? |
| | Q05871 | | P | H | E | R | Saccharomyces cerevisiae | ISOMERASE |
| 0 | 1hnu_2 | A & A | 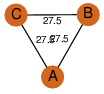 | PROBYES
| D3 | 6 | D3 | 6 | NO |
C3 | 3 | Q05871 | | P | H | E | R | Saccharomyces cerevisiae | ISOMERASE |
| 0 | 1i3c_1 | D & B |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q55169 | | P | H | E | R | Synechocystis sp. | SIGNALING PROTEIN |
| 0 | 1jlk_1 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q55169 | | P | H | E | R | Synechocystis sp. | SIGNALING PROTEIN |
| 0 | 1jxz_1 | A & B | 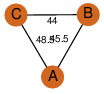 | PROBNOT
| C3 | 3 | C3 | 3 | NO |
C3 | 3 | 419529 | | P | H | E | R | Pseudomonas sp. CBS3 | HYDROLASE |
| 0 | 1k39_1 | A & E |  | NO
| D3 | 6 | D3 | 6 | NA1 |
D3 | 6 | Q05871 | | P | H | E | R | Saccharomyces cerevisiae | ISOMERASE |
| 0 | 1nzy_1 | A & A |  | PROBYES
| D3 | 6 | C3 | 3 | YES |
C3 | 3 | A42560 | | P | H | E | R | Pseudomonas sp. | LYASE |
| 0 | 1nzy_2 | A & A | 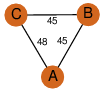 | PROBNOT
| C3 | 3 | C3 | 3 | NO |
C3 | 3 | A42560 | | P | H | E | R | Pseudomonas sp. | LYASE |
| 0 | 1o8u_1 | A & E |  | NO
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | Q93TU6 | | P | H | E | R | RHODOCOCCUS ERYTHROPOLIS | HYDROLASE |
| 0 | 1pjh_1 | A & A |  | NO
| D3 | 6 | D3 | 6 | NA1 |
D3 | 6 | Q05871 | | P | H | E | R | Saccharomyces cerevisiae | ISOMERASE |
| 0 | 1rjm_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 0 | 1rjn_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 0 | 1szo_1 | A & A | 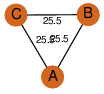 | PROBYES
| C3 | 3 | D3 | 6 | YES |
D3 | 6 | Q93TU6 | | P | H | E | R | Rhodococcus sp. NCIMB 9784 | hydrolase |
| 0 | 1szo_2 | A & F | 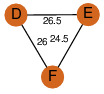 | PROBYES
| C3 | 3 | D3 | 6 | PROBYES2 |
D3 | 6 | Q93TU6 | | P | H | E | R | Rhodococcus sp. NCIMB 9784 | hydrolase |
| 0 | 1szo_3 | A & G |  | PROBYES
| C3 | 3 | D3 | 6 | PROBYES2 |
D3 | 6 | Q93TU6 | | P | H | E | R | Rhodococcus sp. NCIMB 9784 | hydrolase |
| 0 | 1szo_4 | A & J | 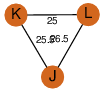 | PROBYES
| C3 | 3 | D3 | 6 | PROBYES2 |
D3 | 6 | Q93TU6 | | P | H | E | R | Rhodococcus sp. NCIMB 9784 | hydrolase |
| 0 | 1szo_5 | A & I |  | PROBNOT
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | Q93TU6 | | P | H | E | R | Rhodococcus sp. NCIMB 9784 | hydrolase |
| 0 | 1szo_6 | A & A |  | PROBNOT
| D3 | 6 | D3 | 6 | NO |
D3 | 6 | Q93TU6 | | P | H | E | R | Rhodococcus sp. NCIMB 9784 | hydrolase |
| 0 | 1wz8_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q5SLS5 | | P | H | E | R | Thermus thermophilus | Lyase |
| 0 | 2a7k_1 | A & C | 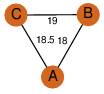 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9XB60 | | P | H | E | R | Pectobacterium carotovorum | Biosynthetic Protein |
| 0 | 2a7k_2 | A & F | 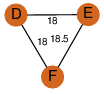 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9XB60 | | P | H | E | R | Pectobacterium carotovorum | Biosynthetic Protein |
| 0 | 2a7k_3 | A & G | 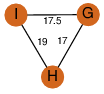 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9XB60 | | P | H | E | R | Pectobacterium carotovorum | Biosynthetic Protein |
| 0 | 2a81_1 | A & A | 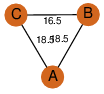 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9XB60 | | P | H | E | R | Pectobacterium carotovorum | Biosynthetic Protein |
| 0 | 2b4a_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9K8I2 | | P | H | E | R | Bacillus halodurans | SIGNALING PROTEIN |
| 0 | 2f6q_1 | A & C | 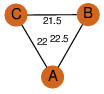 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | O75521 | | P | H | E | R | Homo sapiens | ISOMERASE |
| 0 | 2fbm_1 | A & A | 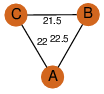 | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q9Y6F8 | | P | H | E | R | Homo sapiens | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2gkg_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | 4063845 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 0 | 2i6f_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1D4U9 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 0 | 2i6f_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1D4U9 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 0 | 2i6f_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q1D4U9 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 0 | 2j5g_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | 2J5G | | P | H | E | R | ANABAENA SP. | HYDROLASE |
| 0 | 2j5g_2 | A & H |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | 2J5G | | P | H | E | R | ANABAENA SP. | HYDROLASE |
| 0 | 2j5s_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | 2J5S | | P | H | E | R | ANABAENA SP. | HYDROLASE |
| 0 | 2nt3_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | O68522 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 0 | 2nt4_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | O68522 | | P | H | E | R | Myxococcus xanthus | SIGNALING PROTEIN |
| 0 | 2pg8_1 | A & E |  | ?
| D3 | 6 | ? | ? | NA1 |
D3 | 6 | Q8KLK7 | | P | H | E | R | Streptomyces toyocaensis | LIGAND BINDING PROTEIN |
| 0 | 2pg8_2 | A & B | 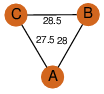 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q8KLK7 | | P | H | E | R | Streptomyces toyocaensis | LIGAND BINDING PROTEIN |
| 0 | 2pln_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O25684 | | P | H | E | R | Helicobacter pylori | SIGNALING PROTEIN |
| 0 | 2q2x_1 | A & A | 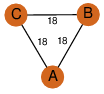 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q6DNE7 | | P | H | E | R | Lyngbya majuscula | LYASE |
| 0 | 2q34_1 | A & B | 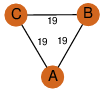 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q6DNE7 | | P | H | E | R | Lyngbya majuscula | LYASE |
| 0 | 2qv0_1 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | P21649 | | P | H | E | R | Klebsiella pneumoniae | TRANSCRIPTION |
| 0 | 2qv0_2 | D & A |  | ?
| C2 | 2 | ? | ? | PROBYES3 |
C2 | 2 | P21649 | | P | H | E | R | Klebsiella pneumoniae | TRANSCRIPTION |
| 0 | 2qzj_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q180B0 | | P | H | E | R | Clostridium difficile | TRANSCRIPTION |
| 0 | 2qzj_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q180B0 | | P | H | E | R | Clostridium difficile | TRANSCRIPTION |
| 0 | 2qzj_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q180B0 | | P | H | E | R | Clostridium difficile | TRANSCRIPTION |
| 0 | 2qzj_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q180B0 | | P | H | E | R | Clostridium difficile | TRANSCRIPTION |
| 0 | 2qzj_5 | D & E |  | ?
| NPS | 1 | ? | ? | ? |
| | Q180B0 | | P | H | E | R | Clostridium difficile | TRANSCRIPTION |
| 0 | 2qzj_6 | D & F |  | ?
| NPS | 1 | ? | ? | ? |
| | Q180B0 | | P | H | E | R | Clostridium difficile | TRANSCRIPTION |
| 0 | 2rdm_1 | D & C |  | ?
| D3 | 6 | ? | ? | ? |
| | A6UIY7 | | P | H | E | R | Sinorhizobium medicae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2rdm_2 | D & A |  | ?
| C3 | 3 | ? | ? | ? |
| | A6UIY7 | | P | H | E | R | Sinorhizobium medicae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2vre_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | 2VRE | | P | H | E | R | HOMO SAPIENS | ISOMERASE |
| 0 | 3cnb_1 | D & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | Q47UF8 | | P | H | E | R | Colwellia psychrerythraea | DNA BINDING PROTEIN |
| 0 | 3cnb_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q47UF8 | | P | H | E | R | Colwellia psychrerythraea | DNA BINDING PROTEIN |
| 0 | 3cnb_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q47UF8 | | P | H | E | R | Colwellia psychrerythraea | DNA BINDING PROTEIN |
| 0 | 3eqz_1 | D & A |  | ?
| D2 | 4 | ? | ? | ? |
| | Q483U6 | | P | H | E | R | Colwellia psychrerythraea | structural genomics, unknown function |
| 0 | 3eqz_2 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q483U6 | | P | H | E | R | Colwellia psychrerythraea | structural genomics, unknown function |
| 0 | 3eqz_3 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q483U6 | | P | H | E | R | Colwellia psychrerythraea | structural genomics, unknown function |
| 0 | 3fdu_1 | A & C | 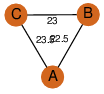 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A3M7S1 | | P | H | E | R | Acinetobacter baumannii | ISOMERASE |
| 0 | 3fdu_2 | A & F |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A3M7S1 | | P | H | E | R | Acinetobacter baumannii | ISOMERASE |
| 0 | 3h0u_1 | A & A |  | ?
| C3 | 3 | ? | ? | YES |
D3 | 6 | Q82Q85 | | P | H | E | R | Streptomyces avermitilis | LYASE |
| 0 | 3h0u_2 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q82Q85 | | P | H | E | R | Streptomyces avermitilis | LYASE |
| 0 | 3he2_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P64014 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 0 | 3hp0_1 | A & A |  | ?
| NPS | 1 | ? | ? | YES |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_2 | A & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_3 | A & C |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_4 | A & D |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_5 | A & E |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_6 | A & F |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_7 | A & E |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3hp0_8 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P40805 | | P | H | E | R | Bacillus subtilis | LYASE |
| 0 | 3ilh_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q11VD8 | | P | H | E | R | Cytophaga hutchinsonii | transcription regulator |
| 0 | 3ilh_2 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q11VD8 | | P | H | E | R | Cytophaga hutchinsonii | transcription regulator |
| 0 | 3isa_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q7W3D6 | | P | H | E | R | Bordetella parapertussis | HYDROLASE |
| 0 | 3kcn_1 | D & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q7UJS6 | | P | H | E | R | Rhodopirellula baltica | LYASE |
| 0 | 3l3s_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5LQZ3 | | P | H | E | R | Ruegeria pomeroyi | ISOMERASE |
| 0 | 3l3s_2 | A & F |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5LQZ3 | | P | H | E | R | Ruegeria pomeroyi | ISOMERASE |
| 0 | 3l3s_3 | A & G |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5LQZ3 | | P | H | E | R | Ruegeria pomeroyi | ISOMERASE |
| 0 | 3l3s_4 | A & L | 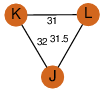 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q5LQZ3 | | P | H | E | R | Ruegeria pomeroyi | ISOMERASE |
| 0 | 3lua_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | A3DK02 | | P | H | E | R | Clostridium thermocellum | Transcription regulator |
| 0 | 3myb_1 | A & A | 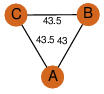 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A0QST4 | | P | H | E | R | Mycobacterium smegmatis | LYASE |
| 0 | 3n0r_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A2S9 | | P | H | E | R | Caulobacter vibrioides | SIGNALING PROTEIN |
| 0 | 3ome_1 | A & C |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | A0R1R4 | | P | H | E | R | Mycobacterium smegmatis | Lyase |
| 0 | 3ot6_1 | A & A |  | ?
| NPS | 1 | ? | ? | YES |
C3 | 3 | Q884M3 | | P | H | E | R | Pseudomonas syringae pv. tomato | ISOMERASE |
| 0 | 3ot6_2 | A & A | 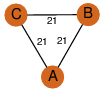 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q884M3 | | P | H | E | R | Pseudomonas syringae pv. tomato | ISOMERASE |
| 0 | 3q1t_1 | A & E |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | A0QIL3 | | P | H | E | R | Mycobacterium avium | LYASE |
| 0 | 3qk8_1 | A & C |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | B2HM22 | | P | H | E | R | Mycobacterium marinum M | LYASE |
| 0 | 3qmj_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B2HD64 | | P | H | E | R | Mycobacterium marinum | LYASE |
| 0 | 3qre_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B2HQD1 | | P | H | E | R | Mycobacterium marinum M | LYASE |
| 0 | 3r6h_1 | A & A | 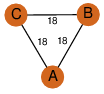 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B2HSG1 | | P | H | E | R | Mycobacterium marinum M | LYASE |
| 0 | 3sll_1 | A & C |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | B1MC45 | | P | H | E | R | Mycobacterium abscessus | ISOMERASE |
| 0 | 3t8a_1 | A & F |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE/LYASE INHIBITOR |
| 0 | 3t8b_1 | A & E |  | ?
| D3 | 6 | ? | ? | PROBYES3 |
D3 | 6 | O06414 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 0 | 3tlf_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q73XB1 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | ISOMERASE |
| 0 | 3tlf_2 | A & A |  | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q73XB1 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | ISOMERASE |
| 0 | 3tlf_3 | A & F |  | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | Q73XB1 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | ISOMERASE |
| 0 | 4dad_1 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q63I75 | | P | H | E | R | Burkholderia pseudomallei | SIGNALING PROTEIN, SIGNAL TRANSDUCTION |
| 0 | 4dn6_1 | D & B |  | ?
| C2 | 2 | ? | ? | ? |
| | Q2T2Y8 | | P | H | E | R | Burkholderia thailandensis | SIGNALING PROTEIN |
| 0 | 4fn7_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P96907 | | P | H | E | R | Mycobacterium tuberculosis | ISOMERASE |
| 0 | 4fn8_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P96907 | | P | H | E | R | Mycobacterium tuberculosis | ISOMERASE |
| 0 | 4fn8_2 | A & F |  | ?
| D3 | 6 | ? | ? | ? |
| | P96907 | | P | H | E | R | Mycobacterium tuberculosis | ISOMERASE |
| 0 | 4fnb_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P96907 | | P | H | E | R | Mycobacterium tuberculosis | ISOMERASE |
| 0 | 4fnd_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P96907 | | P | H | E | R | Mycobacterium tuberculosis | ISOMERASE |
| 0 | 4g97_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q2YRH9 | | P | H | E | R | Brucella melitensis biovar Abortus | SIGNALING PROTEIN |
| 0 | 4hc8_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P96907 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 0 | 4hc8_2 | A & A | 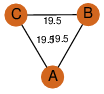 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | P96907 | | P | H | E | R | Mycobacterium tuberculosis | LYASE |
| 0 | 4jcs_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q1LBW6 | | P | H | E | R | Cupriavidus metallidurans | LYASE |
| 0 | 4jot_1 | A & A | 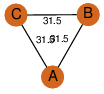 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q9RY37 | | P | H | E | R | Deinococcus radiodurans | LYASE |
| 0 | 4jsb_1 | A & A | 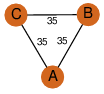 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q47NQ8 | | P | H | E | R | Thermobifida fusca | LYASE |
| 0 | 4jvt_1 | A & C | 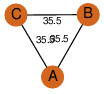 | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q47NQ8 | | P | H | E | R | Thermobifida fusca | LYASE |
| 0 | 4jyj_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q2GB08 | | P | H | E | R | Novosphingobium aromaticivorans | ISOMERASE |
| 0 | 4jyl_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | Q97CA4 | | P | H | E | R | Thermoplasma volcanium | LYASE |
| 0 | 4k29_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | A7IKN6 | | P | H | E | R | Xanthobacter autotrophicus | ISOMERASE |
| 0 | 4k29_2 | A & A |  | ?
| C3 | 3 | ? | ? | PROBYES2 |
D3 | 6 | A7IKN6 | | P | H | E | R | Xanthobacter autotrophicus | ISOMERASE |
| 0 | 4k2n_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q2W7Q6 | | P | H | E | R | Magnetospirillum magneticum | ISOMERASE |
| 0 | 4k3w_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A1U4G2 | | P | H | E | R | Marinobacter aquaeolei | ISOMERASE |
| 0 | 4kd6_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1GB42 | | P | H | E | R | Burkholderia graminis | ISOMERASE |
| 0 | 4kpk_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A8H6D2 | | P | H | E | R | Shewanella pealeana | ISOMERASE |
| 0 | 4lda_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_5 | D & E |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_6 | D & F |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_7 | D & G |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4lda_8 | D & H |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9HW97 | | P | H | E | R | Pseudomonas aeruginosa | TRANSCRIPTION |
| 0 | 4le0_1 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le0_2 | D & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le1_1 | D & A |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le1_2 | D & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le1_3 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le2_1 | D & B |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le2_2 | D & C |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le2_3 | D & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4le2_4 | D & C |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | O34723 | | P | H | E | R | Bacillus subtilis subsp. subtilis | DNA BINDING PROTEIN |
| 0 | 4lk5_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q73WM1 | | P | H | E | R | Mycobacterium avium subsp. paratuberculosis | LYASE |
| 0 | 4lzl_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | J1QDX6 | | P | H | E | R | Streptococcus pneumoniae 2061617 | Transcription |
| 0 | 4mi2_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | B1MJ43 | | P | H | E | R | Mycobacterium abscessus | ISOMERASE |
| 0 | 4ml3_1 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q8DMW5 | | P | H | E | R | Streptococcus pneumoniae | UNKNOWN FUNCTION |
| 0 | 4ml3_2 | D & D |  | ?
| C2 | 2 | ? | ? | ? |
| | Q8DMW5 | | P | H | E | R | Streptococcus pneumoniae | UNKNOWN FUNCTION |
| 0 | 4mld_1 | D & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q8DMW5 | | P | H | E | R | Streptococcus pneumoniae | UNKNOWN FUNCTION |
| 0 | 4mld_2 | D & C |  | ?
| C2 | 2 | ? | ? | ? |
| | Q8DMW5 | | P | H | E | R | Streptococcus pneumoniae | UNKNOWN FUNCTION |
| 0 | 4myr_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q92YL8 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 0 | 4myr_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q92YL8 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 0 | 4myr_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q92YL8 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 0 | 4myr_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q92YL8 | | P | H | E | R | Sinorhizobium meliloti | TRANSCRIPTION |
| 0 | 4n0p_1 | D & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_2 | D & B |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_3 | D & C |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_4 | D & D |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_5 | D & E |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_6 | D & F |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_7 | D & G |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4n0p_8 | D & H |  | ?
| NPS | 1 | ? | ? | ? |
| | Q9A495 | | P | H | E | R | Caulobacter crescentus | SIGNALING PROTEIN |
| 0 | 4og1_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q2GB23 | | P | H | E | R | Novosphingobium aromaticivorans | LYASE |
| 0 | 4olq_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q0C365 | | P | H | E | R | Hyphomonas neptunium | LYASE |
| 0 | 4olq_2 | A & E |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q0C365 | | P | H | E | R | Hyphomonas neptunium | LYASE |
| 0 | 4omr_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | Q47NQ8 | | P | H | E | R | Thermobifida fusca | LYASE |
| 0 | 4qii_1 | A & A |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P9WNP5 | | P | H | E | R | Mycobacterium tuberculosis H37Rv | LYASE |
| 0 | 4qii_2 | A & G |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P9WNP5 | | P | H | E | R | Mycobacterium tuberculosis H37Rv | LYASE |
| 0 | 4qij_1 | A & C |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P9WNP5 | | P | H | E | R | Mycobacterium tuberculosis H37Rv | LYASE |
| 0 | 4qij_2 | A & L |  | ?
| D3 | 6 | ? | ? | NO |
D3 | 6 | P9WNP5 | | P | H | E | R | Mycobacterium tuberculosis H37Rv | LYASE |
| 0 | 4u18_1 | A & B |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | O75521 | | P | H | E | R | Homo sapiens | ISOMERASE |
| 0 | 4u19_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | O75521 | | P | H | E | R | Homo sapiens | ISOMERASE |
| 0 | 4u1a_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | O75521 | | P | H | E | R | Homo sapiens | ISOMERASE |
| 0 | 4wcz_1 | A & A |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A4XEF6 | | P | H | E | R | Novosphingobium aromaticivorans | ISOMERASE |
| 0 | 4wcz_2 | A & F |  | ?
| C3 | 3 | ? | ? | NO |
C3 | 3 | A4XEF6 | | P | H | E | R | Novosphingobium aromaticivorans | ISOMERASE |
| 0 | 4xlt_1 | D & B |  | ?
| C3 | 3 | ? | ? | ? |
| | C6VVW9 | | P | H | E | R | Dyadobacter fermentans (strain ATCC 700827 / DSM 18053 / NS114) | SIGNALING PROTEIN |



































































































































































































































































































































































































































































































































































































































































