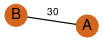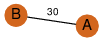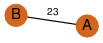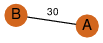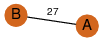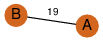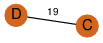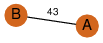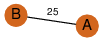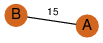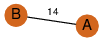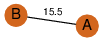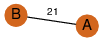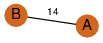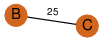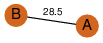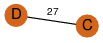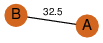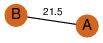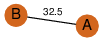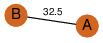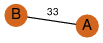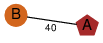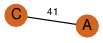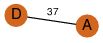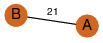to read PiQSi annotation, and roll over the PDB name to read its title.
| %id |
Code |
Chains |
Pic |
Error? |
Sym1 |
Nsub1 |
Sym2 |
Nsub2 |
Error_auto |
Sym_auto |
Nsub_auto |
SProt |
Weight |
Ref |
H |
E |
R |
Org |
Fun |
| 100 | 2nye_2 | A & e |  | ?
| Octa | 48 | ? | ? | PROBYES1 |
C2 | 2 | P12904 | | P | H | E | R | Saccharomyces cerevisiae | PROTEIN BINDING |
| 100 | 2nye_3 | A & C |  | ?
| D4 | 8 | ? | ? | NA1 |
D4 | 8 | P12904 | | P | H | E | R | Saccharomyces cerevisiae | PROTEIN BINDING |
| 100 | 2nye_4 | A & D | 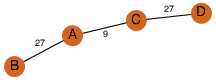 | ?
| C2 | 4 | ? | ? | PROBYES1 |
C2 | 2 | P12904 | | P | H | E | R | Saccharomyces cerevisiae | PROTEIN BINDING |
| 96 | 2nyc_1 | A & A | 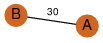 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P12904 | | P | H | E | R | Saccharomyces cerevisiae | PROTEIN BINDING |
| 46 | 2oox_1 | A & G | 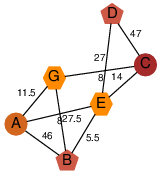 | ?
| C2 | 6 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2oox_2 | A & E | 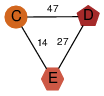 | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2oox_3 | A & G | 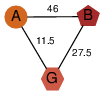 | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2ooy_1 | A & G | 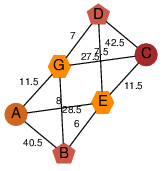 | ?
| C2 | 6 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qr1_1 | A & G | 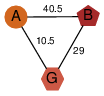 | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qr1_2 | A & E | 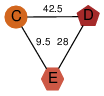 | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qr1_3 | A & G |  | ?
| C2 | 6 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qrc_1 | A & G |  | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qrc_2 | A & E |  | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qrc_3 | A & E |  | ?
| C2 | 6 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qrd_1 | A & G |  | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qrd_2 | A & E |  | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qrd_3 | A & E |  | ?
| C2 | 6 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qre_1 | A & G |  | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qre_2 | A & E | 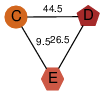 | ?
| NPS | 3 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 46 | 2qre_3 | A & G | 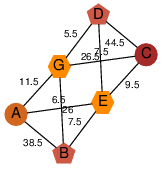 | ?
| C2 | 6 | ? | ? | ? |
| | O74536 | | P | H | E | R | Schizosaccharomyces pombe | TRANSFERASE |
| 37 | 2uv4_1 | A & A | 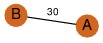 | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | 2UV4 | | P | H | E | R | HOMO SAPIENS | TRANSFERASE |
| 37 | 2uv5_1 | A & B | 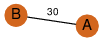 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | 2UV5 | | P | H | E | R | HOMO SAPIENS | TRANSFERASE |
| 37 | 2uv6_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | 2UV6 | | P | H | E | R | HOMO SAPIENS | TRANSFERASE |
| 37 | 2uv7_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | 2UV7 | | P | H | E | R | HOMO SAPIENS | TRANSFERASE |
| 27 | 3sl7_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9C5D0 | | P | H | E | R | Arabidopsis thaliana | MEMBRANE PROTEIN |
| 27 | 4esy_1 | A & A |  | ?
| C2 | 2 | ? | ? | ? |
| | D1CAK9 | | P | H | E | R | Sphaerobacter thermophilus | MEMBRANE PROTEIN |
| 25 | 4gqy_1 | A & A | 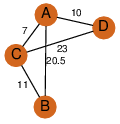 | ?
| C2 | 4 | ? | ? | PROBYES1 |
C2 | 2 | Q9C5D0 | | P | H | E | R | Arabidopsis thaliana | PROTEIN BINDING |
| 25 | 4gqy_2 | A & A | 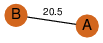 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9C5D0 | | P | H | E | R | Arabidopsis thaliana | PROTEIN BINDING |
| 25 | 4gqy_3 | A & C | 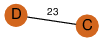 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9C5D0 | | P | H | E | R | Arabidopsis thaliana | PROTEIN BINDING |
| 23 | 3kpb_1 | A & B | 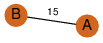 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q57564 | | P | H | E | R | Methanocaldococcus jannaschii | UNKNOWN FUNCTION |
| 23 | 3kpb_2 | A & C | 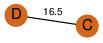 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q57564 | | P | H | E | R | Methanocaldococcus jannaschii | UNKNOWN FUNCTION |
| 23 | 3kpd_1 | A & B | 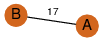 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q57564 | | P | H | E | R | Methanocaldococcus jannaschii | UNKNOWN FUNCTION |
| 23 | 3kpd_2 | A & B | 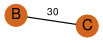 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q57564 | | P | H | E | R | Methanocaldococcus jannaschii | UNKNOWN FUNCTION |
| 23 | 3kpd_3 | A & D | 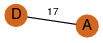 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q57564 | | P | H | E | R | Methanocaldococcus jannaschii | UNKNOWN FUNCTION |
| 22 | 3kpc_1 | A & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q57564 | | P | H | E | R | Methanocaldococcus jannaschii | UNKNOWN FUNCTION |
| 22 | 3lv9_1 | A & A |  | ?
| C2 | 2 | ? | ? | ? |
| | Q183U2 | | P | H | E | R | Clostridium difficile 630 | MEMBRANE PROTEIN |
| 21 | 3fhm_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | A9CIP4 | | P | H | E | R | Agrobacterium tumefaciens str. | structural genomics, unknown function, NUCLEOTIDE-BINDING PROTEIN |
| 21 | 3fhm_2 | A & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | A9CIP4 | | P | H | E | R | Agrobacterium tumefaciens str. | structural genomics, unknown function, NUCLEOTIDE-BINDING PROTEIN |
| 21 | 4gqv_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O23193 | | P | H | E | R | Arabidopsis thaliana | PROTEIN BINDING |
| 21 | 4gqw_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O23193 | | P | H | E | R | Arabidopsis thaliana | PROTEIN BINDING |
| 20 | 2ef7_1 | A & B |  | ?
| D2 | 4 | ? | ? | ? |
| | Q96Y20 | | P | H | E | R | Sulfolobus tokodaii | OXIDOREDUCTASE |
| 20 | 2p9m_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q58332 | | P | H | E | R | Methanocaldococcus jannaschii DSM 2661 | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 20 | 2p9m_2 | A & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q58332 | | P | H | E | R | Methanocaldococcus jannaschii DSM 2661 | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 1o50_1 | A & B |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q9X033 | | P | H | E | R | Thermotoga maritima | TRANSFERASE |
| 0 | 1pbj_1 | A & A |  | ?
| NPS | 1 | ? | ? | ? |
| | O27659 | | P | H | E | R | Methanothermobacter thermautotrophicus str. Delta H | Structural genomics, unknown function |
| 0 | 1pbj_2 | A & B |  | ?
| C2 | 2 | ? | ? | ? |
| | O27659 | | P | H | E | R | Methanothermobacter thermautotrophicus str. Delta H | Structural genomics, unknown function |
| 0 | 1xkf_1 | A & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | O06186 | | P | H | E | R | Mycobacterium tuberculosis | UNKNOWN FUNCTION |
| 0 | 1xkf_2 | A & A |  | ?
| C2 | 2 | ? | ? | PROBYES3 |
C2 | 2 | O06186 | | P | H | E | R | Mycobacterium tuberculosis | UNKNOWN FUNCTION |
| 0 | 1y5h_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O06186 | | P | H | E | R | Mycobacterium tuberculosis | UNKNOWN FUNCTION |
| 0 | 2emq_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q5L147 | | P | H | E | R | Geobacillus kaustophilus | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2j9l_1 | A & A |  | ?
| D3 | 6 | ? | ? | ? |
| | P51795 | | P | H | E | R | HOMO SAPIENS | ION CHANNEL |
| 0 | 2ja3_1 | A & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | 2JA3 | | P | H | E | R | HOMO SAPIENS | TRANSPORT PROTEIN |
| 0 | 2ja3_2 | A & D | 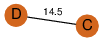 | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | 2JA3 | | P | H | E | R | HOMO SAPIENS | TRANSPORT PROTEIN |
| 0 | 2ja3_3 | A & F | 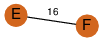 | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | 2JA3 | | P | H | E | R | HOMO SAPIENS | TRANSPORT PROTEIN |
| 0 | 2o16_1 | A & A |  | ?
| NPS | 1 | ? | ? | YES |
C2 | 2 | Q9KTZ3 | | P | H | E | R | Vibrio cholerae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2o16_2 | A & B |  | ?
| NPS | 1 | ? | ? | PROBYES2 |
C2 | 2 | Q9KTZ3 | | P | H | E | R | Vibrio cholerae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2o16_3 | A & A | 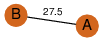 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9KTZ3 | | P | H | E | R | Vibrio cholerae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2o16_4 | A & A | 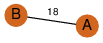 | ?
| C2 | 2 | ? | ? | PROBYES3 |
C2 | 2 | Q9KTZ3 | | P | H | E | R | Vibrio cholerae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2pfi_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P51800 | | P | H | E | R | Homo sapiens | TRANSPORT PROTEIN |
| 0 | 2rc3_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q82SE2 | | P | H | E | R | Nitrosomonas europaea ATCC 19718 | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2rc3_2 | A & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q82SE2 | | P | H | E | R | Nitrosomonas europaea ATCC 19718 | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 2rif_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q8ZVX8 | | P | H | E | R | Pyrobaculum aerophilum | LIGAND-BINDING PROTEIN |
| 0 | 2rif_2 | A & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q8ZVX8 | | P | H | E | R | Pyrobaculum aerophilum | LIGAND-BINDING PROTEIN |
| 0 | 2rif_3 | A & D |  | ?
| C2 | 4 | ? | ? | PROBYES1 |
C2 | 2 | Q8ZVX8 | | P | H | E | R | Pyrobaculum aerophilum | LIGAND-BINDING PROTEIN |
| 0 | 2rih_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q8ZVX8 | | P | H | E | R | Pyrobaculum aerophilum | LIGAND-BINDING PROTEIN |
| 0 | 2yzi_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O57847 | | P | H | E | R | Pyrococcus horikoshii | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 3fv6_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O34994 | | P | H | E | R | Bacillus subtilis | TRANSCRIPTION |
| 0 | 3fwr_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O34994 | | P | H | E | R | Bacillus subtilis | Transcription |
| 0 | 3fws_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | O34994 | | P | H | E | R | Bacillus subtilis | TRANSCRIPTION |
| 0 | 3gby_1 | A & A |  | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q8KDJ9 | | P | H | E | R | Chlorobium tepidum TLS | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 3jtf_1 | A & A | 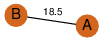 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q7WB69 | | P | H | E | R | Bordetella parapertussis | TRANSPORT PROTEIN |
| 0 | 3k2v_1 | A & A | 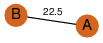 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | A6TEL6 | | P | H | E | R | Klebsiella pneumoniae subsp. pneumoniae | ISOMERASE |
| 0 | 3k6e_1 | A & B | 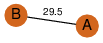 | ?
| C2 | 2 | ? | ? | NO |
C2 | 2 | Q9EUQ9 | | P | H | E | R | Streptococcus pneumoniae | STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
| 0 | 3lhh_1 | A & A |  | ?
| NPS | 1 | ? | ? | ? |
| | Q8EDE1 | | P | H | E | R | Shewanella oneidensis | MEMBRANE PROTEIN |
| 0 | 3lqn_1 | A & A | 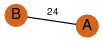 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q81MQ0 | | P | H | E | R | Bacillus anthracis | Structural Genomics, Unknown function |
| 0 | 3nqr_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0A2L3 | | P | H | E | R | Salmonella typhimurium | TRANSPORT PROTEIN |
| 0 | 3nqr_2 | A & C |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | P0A2L3 | | P | H | E | R | Salmonella typhimurium | TRANSPORT PROTEIN |
| 0 | 3oco_1 | A & B |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q04HE1 | | P | H | E | R | Oenococcus oeni | structural genomics, unknown function |
| 0 | 3oi8_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q9K0P8 | | P | H | E | R | Neisseria meningitidis serogroup B | Structural genomics, Unknown function |
| 0 | 4fry_1 | A & A | 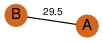 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | B1YXI0 | | P | H | E | R | Burkholderia ambifaria | SIGNALING PROTEIN |
| 0 | 4iy3_1 | A & C | 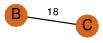 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q6P4Q7 | | P | H | E | R | Homo sapiens | METAL TRANSPORT |
| 0 | 4iy3_2 | A & A | 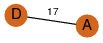 | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q6P4Q7 | | P | H | E | R | Homo sapiens | METAL TRANSPORT |
| 0 | 4noc_1 | A & B | 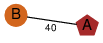 | ?
| C2 | 2 | ? | ? | ? |
| | D2PVT0 | | P | H | E | R | Kribbella flavida | SIGNALING PROTEIN |
| 0 | 4noc_2 | A & A |  | ?
| C2 | 2 | ? | ? | ? |
| | D2PVT0 | | P | H | E | R | Kribbella flavida | SIGNALING PROTEIN |
| 0 | 4noc_3 | A & D |  | ?
| C2 | 2 | ? | ? | ? |
| | D2PVT0 | | P | H | E | R | Kribbella flavida | SIGNALING PROTEIN |
| 0 | 4o9k_1 | A & A |  | ?
| C2 | 2 | ? | ? | PROBNOT |
C2 | 2 | Q60AU8 | | P | H | E | R | Methylococcus capsulatus | ISOMERASE |