Legend
| %id | Code | Chains | Pic | Error? | Sym | Nsub | Sprot | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % seq. id. | PDB Code | query & target | Graph picture | Is the current PDB state judged to be an error? | Symmetry | No. subunits | SwissProt link | No. of amino acids (per chain) | Pubmed ID | Get Homologs | Edit Structure | View structure | Organism | Function |
Protein reference
| Code | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto? | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PDB | PiQSi | |||||||||||||||||
| 1giq_1 |  | ? | NPS | 1 | ? | ? | - | - | - | Q46220 | P | H | E | R | 0 | TOXIN | ||
Homologs
| %id | Code | Chains | Pic | Error? | Sym1 | Nsub1 | Sym2 | Nsub2 | Error_auto | Sym_auto | Nsub_auto | SProt | Weight | Ref | H | E | R | Org | Fun |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 100 | 1giq_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | Q46220 | P | H | E | R | 0 | TOXIN | |||
| 100 | 1gir_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q46220 | P | H | E | R | 0 | TOXIN | |||
| 100 | 3buz_1 | A & A |  | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 100 | 4gy2_1 | A & A |  | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 100 | 4h03_1 | A & A |  | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 100 | 4h0t_1 | A & A |  | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 99 | 4h0v_1 | A & A | 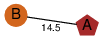 | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 99 | 4h0x_1 | A & A | 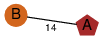 | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 99 | 4h0y_1 | A & A | 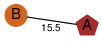 | ? | NPS | 2 | ? | ? | ? | Q46220 | P | H | E | R | Clostridium perfringens | TOXIN/STRUCTURAL PROTEIN | |||
| 85 | 2wn4_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q9KH42 | P | H | E | R | CLOSTRIDIUM DIFFICILE | TRANSFERASE | |||
| 85 | 2wn5_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q9KH42 | P | H | E | R | CLOSTRIDIUM DIFFICILE | TRANSFERASE | |||
| 85 | 2wn6_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q9KH42 | P | H | E | R | CLOSTRIDIUM DIFFICILE | TRANSFERASE | |||
| 85 | 2wn7_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q9KH42 | P | H | E | R | CLOSTRIDIUM DIFFICILE | TRANSFERASE | |||
| 85 | 2wn8_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | Q9KH42 | P | H | E | R | CLOSTRIDIUM DIFFICILE | TRANSFERASE | |||
| 32 | 1qs2_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | 1QS2 | P | H | E | R | Bacillus cereus | TOXIN | |||
| 31 | 1qs1_1 | A & A |  | ? | NPS | 1 | ? | ? | ? | 1QS1 | P | H | E | R | Bacillus cereus | TOXIN | |||
| 31 | 1qs1_2 | A & B |  | ? | NPS | 1 | ? | ? | ? | 1QS1 | P | H | E | R | Bacillus cereus | TOXIN | |||
| 31 | 1qs1_3 | A & C |  | ? | NPS | 1 | ? | ? | ? | 1QS1 | P | H | E | R | Bacillus cereus | TOXIN | |||
| 31 | 1qs1_4 | A & D |  | ? | NPS | 1 | ? | ? | ? | 1QS1 | P | H | E | R | Bacillus cereus | TOXIN |