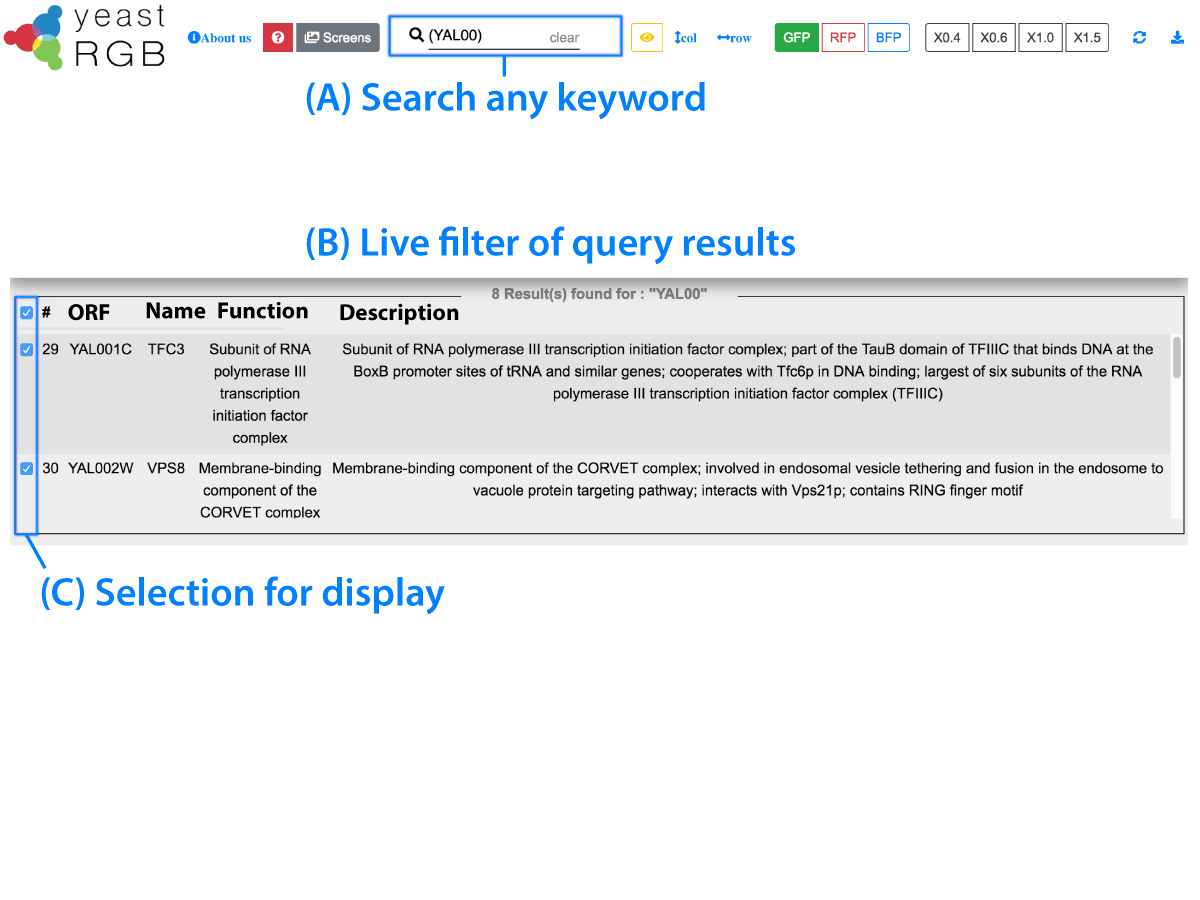
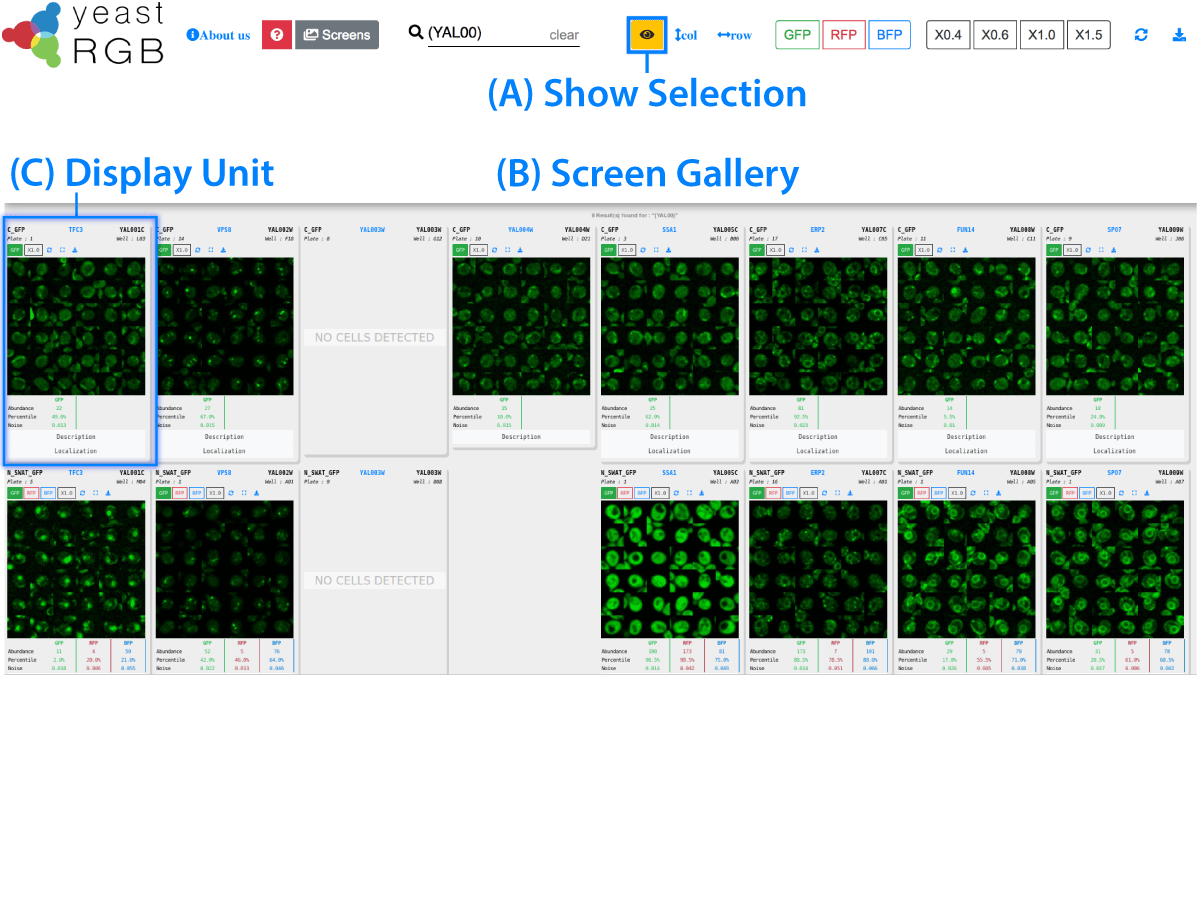
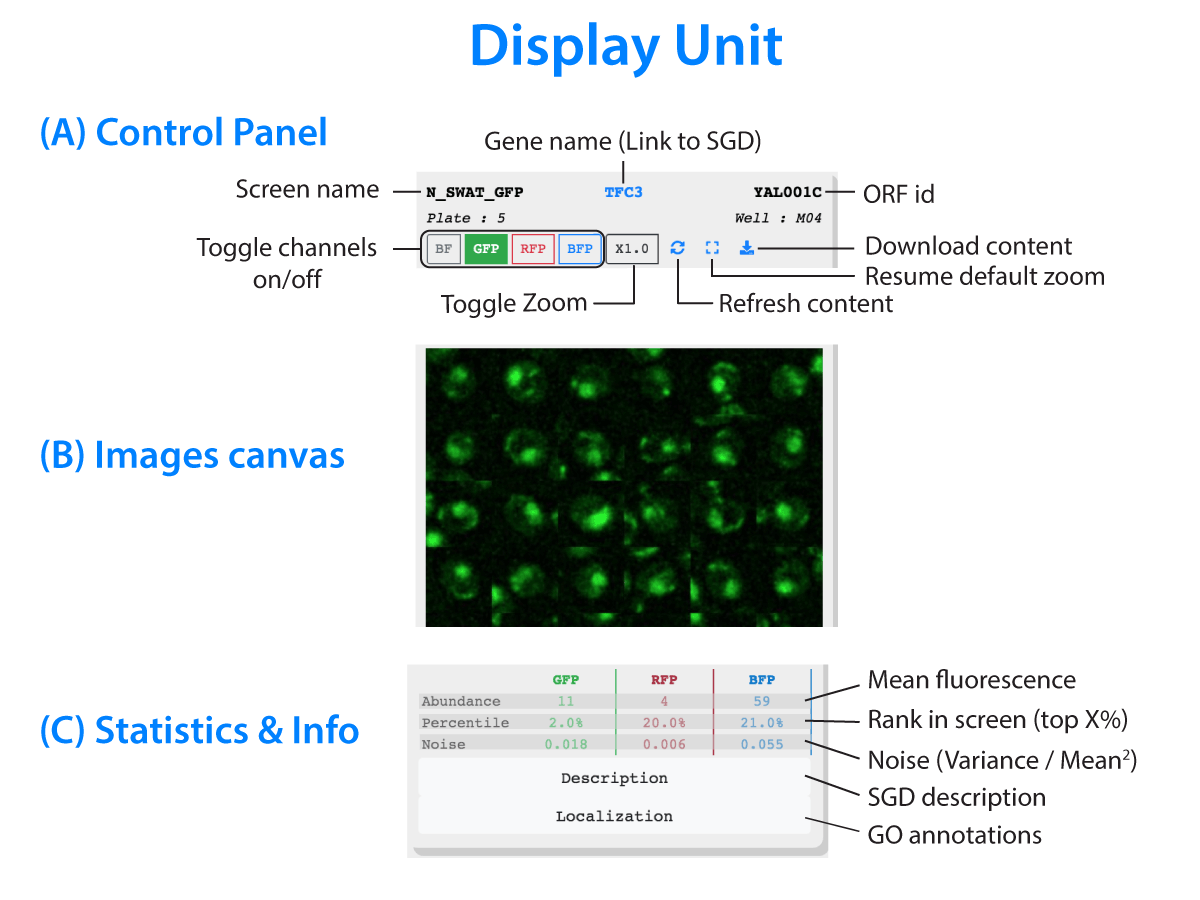
Loading images. Please wait...
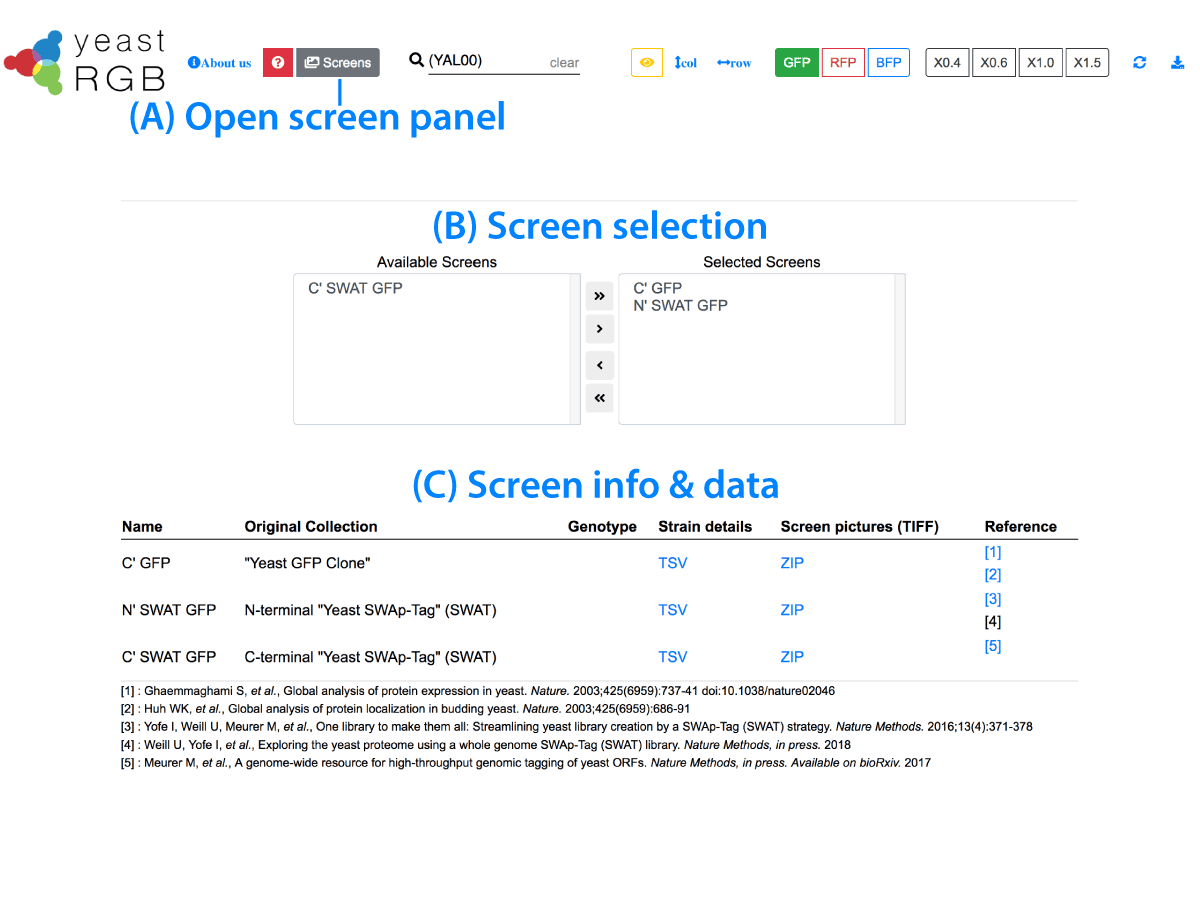
Screen Selection
Available Screens
Selected Screens
| Name | Original Collection | Genotype | Number of Proteins | Strain details | Screen pictures (TIFF) | Reference |
|---|---|---|---|---|---|---|
| C' GFP | "Yeast GFP Clone" | BY4741 background ORF::GFP-HIS3 | 6234 | TSV | NORM (ZIP 5.3GB)RAW (ZIP 5.8GB) |
[1] [2] |
| N' SWAT GFP | N-terminal "Yeast SWAp-Tag" (SWAT) | BY4741 + Y8205 background ORFpromoter::-L1-mCherry-spNOP1-sfGFP-L2::ORF | 5569 | TSV | NORM (ZIP 5.9GB)RAW (ZIP 5.9GB) | [3] [4] |
| C' SWAT GFP | C-terminal "Yeast SWAp-Tag" (SWAT) | BY4741 + Y8205 background ORF:: L3-mNeonGreen-hph-ALG9term-L4crossed withBY4741 background NUP49::mCherry-natNT2 | 5661 | TSV | NORM (ZIP 8.3GB)RAW (ZIP 9.1GB) | [5] [6] |
[1] : Ghaemmaghami S, Huh WK, Bower K, et al., Global analysis of protein expression in yeast. Nature. 2003;425(6959):737-41 [2] : Huh WK, Falvo JV, Gerke LC, et al., Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686-91 [3] : Yofe I, Weill U, Meurer M, et al., One library to make them all: Streamlining yeast library creation by a SWAp-Tag (SWAT) strategy. Nature Methods. 2016;13(4):371-378. [4] : Weill U, Yofe I, Sass E, et al., Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nature Methods 2018;15(8):617-622 [5] : Meurer M, Duan Y, Sass E, et al., Genome-wide C-SWAT library for high-throughput yeast genome tagging. Nature Methods 2018;15(8):598-600 [6] : Meurer M, Duan Y, Sass E, et al., A genome-wide resource for high-throughput genomic tagging of yeast ORFs. BioRxiv 2017;226811